Инструменты и методы ускорения селекции сельскохозяйственных культур

«Tools and Techniques to Accelerate Crop Breeding» из журнала PMC (2024)
Цитирование: Krystal Williams, Mayavan Subramani, Lily W Lofton, Miranda Penney, Antonette Todd, Gulnihal Ozbay и др.
Cоставитель текста: Yarra Rajesh
Аннотация:
Изменение климата и рост численности населения мира продолжают усиливать необходимость увеличения производственных возможностей продовольственных культур, что делает технологические достижения в сельскохозяйственных и crop-исследованиях по-прежнему актуальными. Хотя значительные успехи в селекции растений за последнее столетие способствовали резкому росту урожайности, традиционные методы селекции не обеспечивают достаточного генетического прогресса для удовлетворения будущих потребностей. За последнее десятилетие были разработаны новые методы и инструменты селекции для улучшения сельскохозяйственных культур. Одним из таких достижений является использование скоростной селекции (speed breeding). Скоростная селекция — это применение методов, значительно сокращающих время между поколениями культур, что ускоряет селекцию и исследования. Эти методы быстрого смены поколений помогают ускорить темпы улучшения сельскохозяйственных культур для обеспечения продовольственной безопасности и удовлетворения потребностей растущего населения мира в пище, кормах и волокнах. Скоростная селекция может осуществляться с помощью различных методов, включая оптимизацию условий окружающей среды, геномную селекцию, технологию CRISPR-Cas9 и эпигеномные инструменты. Данный обзор направлен на обсуждение этих значительных достижений в технологиях и методах селекции растений, которые могут значительно улучшить способность селекционеров быстро создавать жизненно важные сорта.
Ключевые слова: скоростная селекция, CRISPR-Cas9, микроорганизмы, геномная селекция, эпигеномика, улучшение сельскохозяйственных культур
1. Введение
По оценкам, к 2050 году население мира достигнет 10 миллиардов человек [1]. Поскольку соотношение производителей продовольствия к его потребителям продолжает значительно снижаться, растет спрос на большее количество сельскохозяйственных культур, выращиваемых на меньших площадях и с меньшими трудозатратами. Этого нельзя достичь только с помощью существующих сортов и методов ведения сельского хозяйства [2]. Текущая продуктивность сельскохозяйственных культур недостаточна и должна быть удвоена, чтобы удовлетворить спрос на растительную продукцию к 2050 году. Потеря разнообразия современных культур из-за эффекта «бутылочного горлышка» доместикации, длительные сроки вывода новых высокопродуктивных сортов и неоптимальное управление сельскохозяйственными практиками требуют новых технологий и подходов для ускоренной селекции культур [2,3].
В основе термин «селекция растений» означает усилия человека по повышению продуктивности культур, что включает отбор желаемых признаков у растительных вариантов для создания более эффективных в производстве культур [4]. Этот процесс основан на использовании генетической изменчивости внутри вида и чаще всего осуществляется с помощью таких методов, как искусственное опыление, культура зародышей, удвоение хромосом и гибридизация [5]. Урожайность, качество, устойчивость к неблагоприятным условиям, устойчивость к вредителям, толерантность к абиотическим стрессам и простота переработки — это распространенные признаки, которые стремятся улучшить селекционеры [4], поскольку эти факторы в значительной степени определяют успех культуры и доступность продовольствия.
Несмотря на их ранее успешное применение в ускорении производства сельскохозяйственных культур, многие существующие методы селекции оказались неэффективными из-за изменения климата и роста населения. Кроме того, хотя окончательное влияние изменения климата на общество трудно предсказать, оно по-прежнему считается серьезной угрозой продовольственной безопасности [6]. Согласно прогнозам, при обычном сценарии роста населения к 2080 году еще 120 миллионов человек будут страдать от недоедания [7,8], и это отчасти связано с нехваткой продовольствия из-за последовательных сезонов неурожаев.
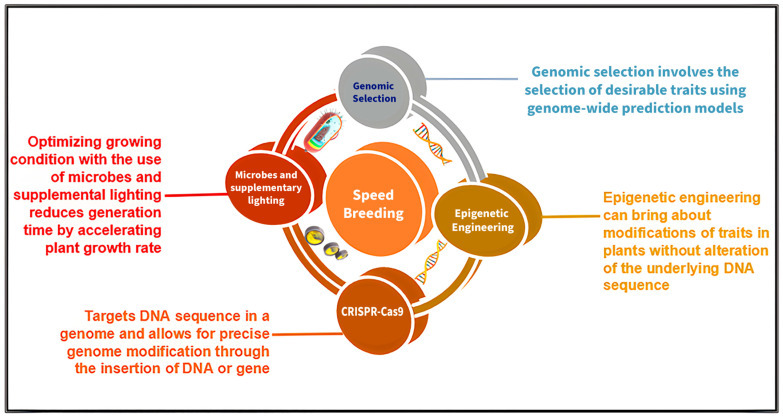
Скоростная селекция — это метод, сокращающий селекционный цикл, что позволяет ускорить смену поколений и тем самым интенсифицировать исследования в области селекции растений [9,10]. Скоростная селекция может осуществляться различными способами, включая увеличение ежедневного освещения растений и ранний сбор семян для сокращения времени смены поколений у нейтральных к длине дня культур [10]. Скоростная селекция в сочетании с другими инструментами и технологиями селекции может оказаться полезной и эффективной для улучшения сельскохозяйственных культур (Рисунок 1). В этом обзоре обсуждаются различные инструменты и методы скоростной селекции. Каждый раздел включает краткое введение о технологиях и методах, а также их применение в скоростной селекции для ускоренного улучшения культур.
Доступ к данным других исследований через научные базы данных крайне важен для изучения скоростной селекции, поскольку это способствует расширению как частоты использования, так и целей применения этих методов. Кроме того, поскольку научные базы данных могут повысить экономическую эффективность, позволяя ученым избегать дорогостоящих ошибок и в целом способствуя созданию ценности в научном сообществе [11], в этом обзоре также приводятся примеры исследований, в которых обсуждаемые технологии уже применялись в селекции растений.
Цитирование: Krystal Williams, Mayavan Subramani, Lily W Lofton, Miranda Penney, Antonette Todd, Gulnihal Ozbay и др.
Cоставитель текста: Yarra Rajesh
Аннотация:
Изменение климата и рост численности населения мира продолжают усиливать необходимость увеличения производственных возможностей продовольственных культур, что делает технологические достижения в сельскохозяйственных и crop-исследованиях по-прежнему актуальными. Хотя значительные успехи в селекции растений за последнее столетие способствовали резкому росту урожайности, традиционные методы селекции не обеспечивают достаточного генетического прогресса для удовлетворения будущих потребностей. За последнее десятилетие были разработаны новые методы и инструменты селекции для улучшения сельскохозяйственных культур. Одним из таких достижений является использование скоростной селекции (speed breeding). Скоростная селекция — это применение методов, значительно сокращающих время между поколениями культур, что ускоряет селекцию и исследования. Эти методы быстрого смены поколений помогают ускорить темпы улучшения сельскохозяйственных культур для обеспечения продовольственной безопасности и удовлетворения потребностей растущего населения мира в пище, кормах и волокнах. Скоростная селекция может осуществляться с помощью различных методов, включая оптимизацию условий окружающей среды, геномную селекцию, технологию CRISPR-Cas9 и эпигеномные инструменты. Данный обзор направлен на обсуждение этих значительных достижений в технологиях и методах селекции растений, которые могут значительно улучшить способность селекционеров быстро создавать жизненно важные сорта.
Ключевые слова: скоростная селекция, CRISPR-Cas9, микроорганизмы, геномная селекция, эпигеномика, улучшение сельскохозяйственных культур
1. Введение
По оценкам, к 2050 году население мира достигнет 10 миллиардов человек [1]. Поскольку соотношение производителей продовольствия к его потребителям продолжает значительно снижаться, растет спрос на большее количество сельскохозяйственных культур, выращиваемых на меньших площадях и с меньшими трудозатратами. Этого нельзя достичь только с помощью существующих сортов и методов ведения сельского хозяйства [2]. Текущая продуктивность сельскохозяйственных культур недостаточна и должна быть удвоена, чтобы удовлетворить спрос на растительную продукцию к 2050 году. Потеря разнообразия современных культур из-за эффекта «бутылочного горлышка» доместикации, длительные сроки вывода новых высокопродуктивных сортов и неоптимальное управление сельскохозяйственными практиками требуют новых технологий и подходов для ускоренной селекции культур [2,3].
В основе термин «селекция растений» означает усилия человека по повышению продуктивности культур, что включает отбор желаемых признаков у растительных вариантов для создания более эффективных в производстве культур [4]. Этот процесс основан на использовании генетической изменчивости внутри вида и чаще всего осуществляется с помощью таких методов, как искусственное опыление, культура зародышей, удвоение хромосом и гибридизация [5]. Урожайность, качество, устойчивость к неблагоприятным условиям, устойчивость к вредителям, толерантность к абиотическим стрессам и простота переработки — это распространенные признаки, которые стремятся улучшить селекционеры [4], поскольку эти факторы в значительной степени определяют успех культуры и доступность продовольствия.
Несмотря на их ранее успешное применение в ускорении производства сельскохозяйственных культур, многие существующие методы селекции оказались неэффективными из-за изменения климата и роста населения. Кроме того, хотя окончательное влияние изменения климата на общество трудно предсказать, оно по-прежнему считается серьезной угрозой продовольственной безопасности [6]. Согласно прогнозам, при обычном сценарии роста населения к 2080 году еще 120 миллионов человек будут страдать от недоедания [7,8], и это отчасти связано с нехваткой продовольствия из-за последовательных сезонов неурожаев.
Скоростная селекция — это метод, сокращающий селекционный цикл, что позволяет ускорить смену поколений и тем самым интенсифицировать исследования в области селекции растений [9,10]. Скоростная селекция может осуществляться различными способами, включая увеличение ежедневного освещения растений и ранний сбор семян для сокращения времени смены поколений у нейтральных к длине дня культур [10]. Скоростная селекция в сочетании с другими инструментами и технологиями селекции может оказаться полезной и эффективной для улучшения сельскохозяйственных культур (Рисунок 1). В этом обзоре обсуждаются различные инструменты и методы скоростной селекции. Каждый раздел включает краткое введение о технологиях и методах, а также их применение в скоростной селекции для ускоренного улучшения культур.
Доступ к данным других исследований через научные базы данных крайне важен для изучения скоростной селекции, поскольку это способствует расширению как частоты использования, так и целей применения этих методов. Кроме того, поскольку научные базы данных могут повысить экономическую эффективность, позволяя ученым избегать дорогостоящих ошибок и в целом способствуя созданию ценности в научном сообществе [11], в этом обзоре также приводятся примеры исследований, в которых обсуждаемые технологии уже применялись в селекции растений.
2. Оптимизация условий выращивания
Многие сельскохозяйственные культуры в естественных условиях могут давать лишь одно-два поколения в год, даже при использовании зимних рассадников, которые позволяют ускорить селекцию за счет получения дополнительных поколений в год [12]. В результате на создание новых сортов уходит несколько лет. Чтобы сократить время смены поколений, растения можно выращивать в оптимальных условиях, ускоряя их рост и стимулируя раннее цветение с помощью физиологического стресса [10,13]. Использование искусственного освещения и микроорганизмов — два метода, которые могут ускорить этот процесс, сокращая время выведения новых стабильных линий, пригодных для агрономической оценки.
2.1. Дополнительное освещение
Рост и развитие растений зависят от нескольких семейств фоторецепторов, которые помогают им реагировать на внешние факторы среды, такие как свет [14]. Свет влияет на молекулярные механизмы развития растений. Изменения в профилях экспрессии генов в ответ на свет были зафиксированы у *Arabidopsis* и риса [15]. Реакция растений на свет варьируется в зависимости от длины волны. Поскольку свет является необходимым источником энергии для жизни растений, правильный баланс световой энергии улучшает их развитие в целом.
Свет может поступать к растениям естественным образом от солнца или от искусственных источников. Использование дополнительного освещения в контролируемых условиях крайне важно для круглогодичного выращивания здоровых растений. Одно из главных преимуществ этого подхода — нивелирование сезонных колебаний солнечной радиации и обеспечение растений необходимой суточной нормой освещения (DLI) [16]. Существует несколько видов дополнительного освещения, включая натриевые лампы высокого давления (HPS), светодиоды (LED), керамические металлогалогенные и люминесцентные лампы [17,18]. Однако в последние годы светодиоды стали предпочтительным источником света благодаря высокой энергоэффективности, низкой стоимости и широкой доступности.
Недавно Стефаньски и соавт. [19] провели исследование, сравнивая разные типы дополнительного освещения при выращивании зерновых культур, и пришли к выводу, что светодиодные лампы более энергоэффективны по сравнению с HPS и газоразрядными лампами высокой интенсивности (HID) в селекции овса (*Avena sativa* L.), ячменя (*Hordeum vulgare* L.) и пшеницы (*Triticum aestivum* L.) (Рисунок 2).
Многие сельскохозяйственные культуры в естественных условиях могут давать лишь одно-два поколения в год, даже при использовании зимних рассадников, которые позволяют ускорить селекцию за счет получения дополнительных поколений в год [12]. В результате на создание новых сортов уходит несколько лет. Чтобы сократить время смены поколений, растения можно выращивать в оптимальных условиях, ускоряя их рост и стимулируя раннее цветение с помощью физиологического стресса [10,13]. Использование искусственного освещения и микроорганизмов — два метода, которые могут ускорить этот процесс, сокращая время выведения новых стабильных линий, пригодных для агрономической оценки.
2.1. Дополнительное освещение
Рост и развитие растений зависят от нескольких семейств фоторецепторов, которые помогают им реагировать на внешние факторы среды, такие как свет [14]. Свет влияет на молекулярные механизмы развития растений. Изменения в профилях экспрессии генов в ответ на свет были зафиксированы у *Arabidopsis* и риса [15]. Реакция растений на свет варьируется в зависимости от длины волны. Поскольку свет является необходимым источником энергии для жизни растений, правильный баланс световой энергии улучшает их развитие в целом.
Свет может поступать к растениям естественным образом от солнца или от искусственных источников. Использование дополнительного освещения в контролируемых условиях крайне важно для круглогодичного выращивания здоровых растений. Одно из главных преимуществ этого подхода — нивелирование сезонных колебаний солнечной радиации и обеспечение растений необходимой суточной нормой освещения (DLI) [16]. Существует несколько видов дополнительного освещения, включая натриевые лампы высокого давления (HPS), светодиоды (LED), керамические металлогалогенные и люминесцентные лампы [17,18]. Однако в последние годы светодиоды стали предпочтительным источником света благодаря высокой энергоэффективности, низкой стоимости и широкой доступности.
Недавно Стефаньски и соавт. [19] провели исследование, сравнивая разные типы дополнительного освещения при выращивании зерновых культур, и пришли к выводу, что светодиодные лампы более энергоэффективны по сравнению с HPS и газоразрядными лампами высокой интенсивности (HID) в селекции овса (*Avena sativa* L.), ячменя (*Hordeum vulgare* L.) и пшеницы (*Triticum aestivum* L.) (Рисунок 2).
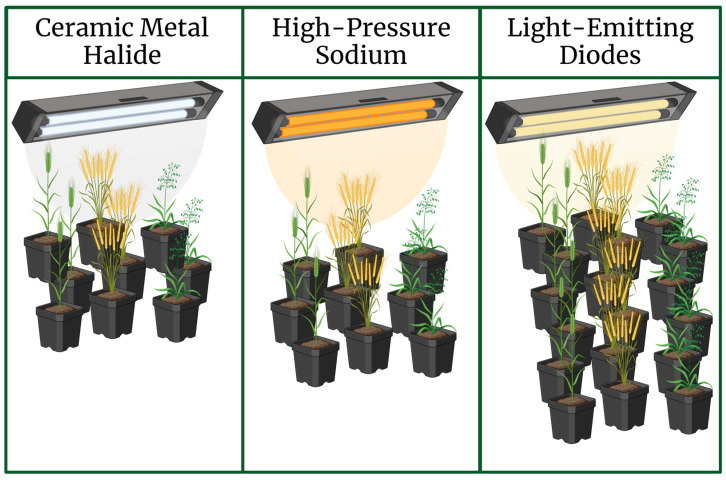
Сравнение влияния различных типов дополнительного освещения на скорость смены поколений.
Помимо общей энергоэффективности и эксплуатационных затрат, светодиоды (LED) превосходят керамические металлогалогенные и натриевые лампы высокого давления по количеству поколений в год у ячменя, пшеницы и овса [19].
Помимо общей энергоэффективности и эксплуатационных затрат, светодиоды (LED) превосходят керамические металлогалогенные и натриевые лампы высокого давления по количеству поколений в год у ячменя, пшеницы и овса [19].
Разработки в области светодиодов и их эффективность для роста и развития сельскохозяйственных культур [20] открыли новые возможности для эффективного растениеводства и выращивания растений в контролируемых условиях [21,22]. Интенсивность и качество светодиодного освещения могут улучшать метаболизм, урожайность и качество культур [23]. Светодиодные системы обычно потребляют меньше электроэнергии и максимизируют эффективность поглощения фотонов, что крайне важно для ускорения роста растений и, как следствие, увеличения скорости смены поколений у некоторых культур [12] (Рисунок 2).
Методы выращивания в помещении часто используются селекционерами для ускорения селекционного процесса и избегания неблагоприятных сезонных колебаний, характерных для открытого грунта [23]. Применение светодиодов в условиях закрытого грунта снижает общие производственные затраты благодаря их высокой эффективности, низким требованиям к обслуживанию и долговечности [24]. Использование светодиодов распространяется на системы контролируемого земледелия (CEA), включая теплицы, климатические камеры и вертикальные фермы [25].
Скоростная селекция может применяться независимо от генотипа и ускорять смену поколений за счет раннего сбора семян в контролируемой среде с оптимальной интенсивностью и качеством светодиодного освещения. Это уже успешно реализовано для пшеницы, ячменя, нута, гороха, рапса [26,27] и некоторых малораспространенных культур [28]. Например, система скоростной селекции с увеличенным фотопериодом (22 часа в сутки) позволила получать до шести поколений яровой пшеницы и ярового ячменя в год [27], а комбинация скоростной селекции с ускоренной яровизацией дала возможность получать до пяти поколений озимого ячменя и озимой пшеницы [29].
Светодиоды также могут использоваться в овощеводстве и цветоводстве для повышения урожайности, улучшения качества продукции до и после сбора, увеличения содержания фитохимических и питательных веществ [25]. Скоростная селекция культур короткого дня, таких как соя, рис и амарант, позволила получать несколько поколений в год за счет регулирования интенсивности и спектра светодиодного освещения, что ускоряет цветение и созревание. Таким образом, интеллектуальные системы освещения, включая режимы, адаптированные под конкретные культуры, будут играть ключевую роль в увеличении производства на городских фермах, помогая решать проблемы продовольственной безопасности растущего городского населения за счет быстрого доступа к свежим продуктам [30].
В рамках инициативы Фонда Билла и Мелинды Гейтс Ли Хики и его исследовательская группа в сотрудничестве с Международным институтом исследования сельскохозяйственных культур для полузасушливых тропиков (ICRISAT) оценили возможность скоростной селекции культур короткого дня (просо, сорго, голубиный горох) путем оптимизации методов для ускоренного цветения и быстрой смены поколений.
Интеграция дополнительного освещения в контролируемых условиях с высокопроизводительным генотипированием, геномной селекцией и ускоренным созданием улучшенных сортов открывает новые перспективы для селекционных программ [2]. Быстрый рост и раннее цветение растений позволяют получать новое поколение за более короткий срок, что облегчает изучение мутантов и фенотипирование взрослых растений, ускоряя весь селекционный процесс.
2.2. Использование микроорганизмов для стимуляции роста растений
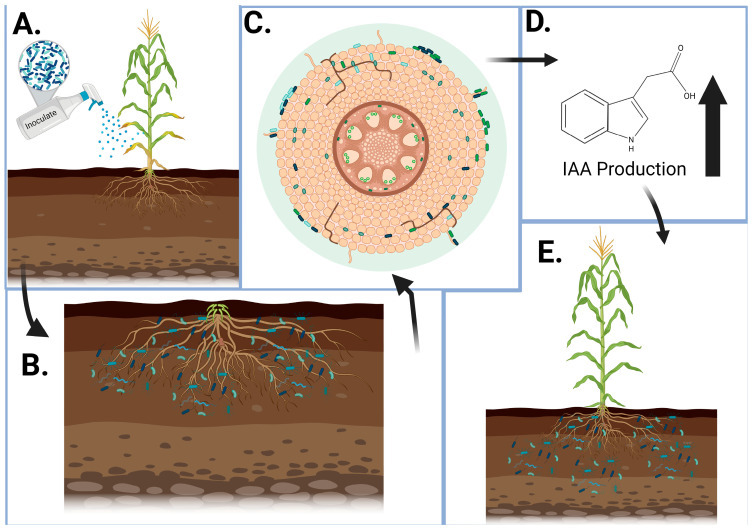
Еще один метод скоростной селекции — применение микроорганизмов. Тысячи видов микробов образуют сложные ассоциации с растениями, которые могут быть полезны для их здоровья [31]. Растения способны формировать свой ризосферный микробиом за счет выделения корневых экссудатов, которые стимулируют рост различных микробных популяций. Как правило, растения одного вида имеют сходный микробный состав [31].
Структурные и функциональные особенности корней влияют на способность растений формировать связи с микроорганизмами, что определяет их потенциал в усвоении питательных веществ [32]. Применение микробов для стимуляции роста растений может сократить время смены поколений, поскольку некоторые виды микроорганизмов регулируют гормональный баланс и вырабатывают фитогормоны, ускоряющие рост [33].
Примеры полезных микроорганизмов:
- Ризобактерии, способствующие росту растений (PGPR), и микоризные грибы колонизируют корни и изменяют их архитектуру. PGPR продуцируют фитогормоны, усиливающие ветвление корней и образование корневых волосков, что улучшает питание и общую физиологию растения [34].
- Микоризные грибы помогают solubilize питательные вещества и повышают устойчивость к патогенам [33]. Например, Glomus mosseae и Rhizophagus irregularis показали биозащитный эффект против возбудителя корневой гнили Aphanomyces euteiches у зернобобовых культур [35].
Бактерии также играют важную роль в защите растений. Например:
- Род Paenibacillus обладает антагонистической активностью против фитопатогенов [36].
- Штаммы Rhizobium leguminosarum снижают заболеваемость корневыми гнилями (например, питиозом) у чечевицы и гороха [37].
Эндофиты (симбиотические микроорганизмы, живущие в тканях растений) могут ускорять рост растений. Например, они стимулировали выработку индол-3-уксусной кислоты (ИУК) у Pulicaria incisa, что относится к классу ауксинов — ключевых гормонов, регулирующих процессы роста, включая деление и удлинение клеток, а также реакции на абиотические и биотические стрессы [38,39].
Как подчеркивалось ранее, полезные микроорганизмы играют важную роль в выращивании здоровых растений (Рисунок 3), что крайне важно для сокращения времени смены поколений в скоростной селекции.
Методы выращивания в помещении часто используются селекционерами для ускорения селекционного процесса и избегания неблагоприятных сезонных колебаний, характерных для открытого грунта [23]. Применение светодиодов в условиях закрытого грунта снижает общие производственные затраты благодаря их высокой эффективности, низким требованиям к обслуживанию и долговечности [24]. Использование светодиодов распространяется на системы контролируемого земледелия (CEA), включая теплицы, климатические камеры и вертикальные фермы [25].
Скоростная селекция может применяться независимо от генотипа и ускорять смену поколений за счет раннего сбора семян в контролируемой среде с оптимальной интенсивностью и качеством светодиодного освещения. Это уже успешно реализовано для пшеницы, ячменя, нута, гороха, рапса [26,27] и некоторых малораспространенных культур [28]. Например, система скоростной селекции с увеличенным фотопериодом (22 часа в сутки) позволила получать до шести поколений яровой пшеницы и ярового ячменя в год [27], а комбинация скоростной селекции с ускоренной яровизацией дала возможность получать до пяти поколений озимого ячменя и озимой пшеницы [29].
Светодиоды также могут использоваться в овощеводстве и цветоводстве для повышения урожайности, улучшения качества продукции до и после сбора, увеличения содержания фитохимических и питательных веществ [25]. Скоростная селекция культур короткого дня, таких как соя, рис и амарант, позволила получать несколько поколений в год за счет регулирования интенсивности и спектра светодиодного освещения, что ускоряет цветение и созревание. Таким образом, интеллектуальные системы освещения, включая режимы, адаптированные под конкретные культуры, будут играть ключевую роль в увеличении производства на городских фермах, помогая решать проблемы продовольственной безопасности растущего городского населения за счет быстрого доступа к свежим продуктам [30].
В рамках инициативы Фонда Билла и Мелинды Гейтс Ли Хики и его исследовательская группа в сотрудничестве с Международным институтом исследования сельскохозяйственных культур для полузасушливых тропиков (ICRISAT) оценили возможность скоростной селекции культур короткого дня (просо, сорго, голубиный горох) путем оптимизации методов для ускоренного цветения и быстрой смены поколений.
Интеграция дополнительного освещения в контролируемых условиях с высокопроизводительным генотипированием, геномной селекцией и ускоренным созданием улучшенных сортов открывает новые перспективы для селекционных программ [2]. Быстрый рост и раннее цветение растений позволяют получать новое поколение за более короткий срок, что облегчает изучение мутантов и фенотипирование взрослых растений, ускоряя весь селекционный процесс.
2.2. Использование микроорганизмов для стимуляции роста растений
Еще один метод скоростной селекции — применение микроорганизмов. Тысячи видов микробов образуют сложные ассоциации с растениями, которые могут быть полезны для их здоровья [31]. Растения способны формировать свой ризосферный микробиом за счет выделения корневых экссудатов, которые стимулируют рост различных микробных популяций. Как правило, растения одного вида имеют сходный микробный состав [31].
Структурные и функциональные особенности корней влияют на способность растений формировать связи с микроорганизмами, что определяет их потенциал в усвоении питательных веществ [32]. Применение микробов для стимуляции роста растений может сократить время смены поколений, поскольку некоторые виды микроорганизмов регулируют гормональный баланс и вырабатывают фитогормоны, ускоряющие рост [33].
Примеры полезных микроорганизмов:
- Ризобактерии, способствующие росту растений (PGPR), и микоризные грибы колонизируют корни и изменяют их архитектуру. PGPR продуцируют фитогормоны, усиливающие ветвление корней и образование корневых волосков, что улучшает питание и общую физиологию растения [34].
- Микоризные грибы помогают solubilize питательные вещества и повышают устойчивость к патогенам [33]. Например, Glomus mosseae и Rhizophagus irregularis показали биозащитный эффект против возбудителя корневой гнили Aphanomyces euteiches у зернобобовых культур [35].
Бактерии также играют важную роль в защите растений. Например:
- Род Paenibacillus обладает антагонистической активностью против фитопатогенов [36].
- Штаммы Rhizobium leguminosarum снижают заболеваемость корневыми гнилями (например, питиозом) у чечевицы и гороха [37].
Эндофиты (симбиотические микроорганизмы, живущие в тканях растений) могут ускорять рост растений. Например, они стимулировали выработку индол-3-уксусной кислоты (ИУК) у Pulicaria incisa, что относится к классу ауксинов — ключевых гормонов, регулирующих процессы роста, включая деление и удлинение клеток, а также реакции на абиотические и биотические стрессы [38,39].
Как подчеркивалось ранее, полезные микроорганизмы играют важную роль в выращивании здоровых растений (Рисунок 3), что крайне важно для сокращения времени смены поколений в скоростной селекции.
3. CRISPR-Cas9
Многие прокариоты, включая бактерии и археи, развили сложные иммунные системы, кодируемые кластерами регулярно расположенных коротких палиндромных повторов (CRISPR) и ассоциированными с CRISPR генами (Cas) [40,41]. Эта система CRISPR-Cas представляет собой РНК-направленную адаптивную иммунную систему, обеспечивающую защиту от инфекций бактериофагами и переноса плазмид [41].
При заражении бактериофагом фрагменты вирусной ДНК интегрируются в виде новых спейсеров в CRISPR-массив (короткие идентичные повторы, разделенные уникальными спейсерами) хромосомы хозяина. Эта интеграция чужеродной ДНК создает генетическую запись или "иммунную память" о прошлых инфекциях, позволяя хозяину защищаться от последующих атак того же патогена [41,42].
Транскрипция и ферментативная обработка CRISPR-массива приводят к образованию зрелых CRISPR-РНК (crРНК). Каждая crРНК содержит новый CRISPR-спейсер (последовательность, комплементарную чужеродному генетическому элементу) на 5'-конце и часть повтора CRISPR на 3'-конце [41]. Система CRISPR-Cas включает транскрипцию CRISPR-локусов и сопутствующих генов Cas, кодирующих белки с нуклеазными и хеликазными доменами [42]. Эти белки используют crРНК для распознавания и инактивации чужеродных РНК и ДНК [40,43].
При повторном заражении crРНК объединяются с Cas-белками, формируя эффекторный комплекс [44]. Спейсерная последовательность в crРНК гибридизуется с комплементарной мишенью (например, вирусной ДНК или плазмидой), что позволяет комплексу распознать чужеродный генетический элемент [41,44]. Это взаимодействие запускает специфичное расщепление мишени Cas-нуклеазами [41], предотвращая распространение чужеродных нуклеиновых кислот.
3.1. Технология CRISPR-Cas9
Системы CRISPR-Cas делятся на шесть типов (I-VI), относящихся к двум классам, которые используют разные механизмы распознавания и расщепления мишеней [41]:
- Класс 1 (типы I, III, IV): многобелковые эффекторные комплексы crРНК
- Класс 2 (типы II, V, VI): одноцепочечные эффекторные комплексы crРНК
Типы I, II и V нацелены на ДНК, тогда как типы III и VI — на РНК [46]. В системах I и III для распознавания и расщепления мишени используется комплекс из нескольких Cas-белков [41,42]. В отличие от них, система типа II использует единственный белок — эндонуклеазу Cas9 — для специфичного расщепления ДНК [41].
Ключевым элементом является мотив PAM (protospacer-adjacent motif) — короткая последовательность, прилегающая к мишени и необходимая для ее распознавания большинством CRISPR-Cas систем, особенно типа II [41,42].
В системе CRISPR-Cas9 типа II:
1. Малая некодирующая tracrРНК связывается с повтором в crРНК [41]
2. Образуется дуплекс crРНК-tracrРНК, направляющий Cas9 к мишени
3. Cas9 расщепляет ДНК, содержащую комплементарный спейсеру участок и соседний PAM
Изменяя последовательность спейсера в guideРНК (гидРНК), можно запрограммировать систему на распознавание практически любого участка генома. Двухцепочечный разрыв затем репарируется:
- Случайно (с образованием инделов)
- Или направленно (с использованием матрицы для гомологичной рекомбинации)
Это позволяет проводить точные геномные модификации, включая вставку целевых генов (Рисунок 4).
Многие прокариоты, включая бактерии и археи, развили сложные иммунные системы, кодируемые кластерами регулярно расположенных коротких палиндромных повторов (CRISPR) и ассоциированными с CRISPR генами (Cas) [40,41]. Эта система CRISPR-Cas представляет собой РНК-направленную адаптивную иммунную систему, обеспечивающую защиту от инфекций бактериофагами и переноса плазмид [41].
При заражении бактериофагом фрагменты вирусной ДНК интегрируются в виде новых спейсеров в CRISPR-массив (короткие идентичные повторы, разделенные уникальными спейсерами) хромосомы хозяина. Эта интеграция чужеродной ДНК создает генетическую запись или "иммунную память" о прошлых инфекциях, позволяя хозяину защищаться от последующих атак того же патогена [41,42].
Транскрипция и ферментативная обработка CRISPR-массива приводят к образованию зрелых CRISPR-РНК (crРНК). Каждая crРНК содержит новый CRISPR-спейсер (последовательность, комплементарную чужеродному генетическому элементу) на 5'-конце и часть повтора CRISPR на 3'-конце [41]. Система CRISPR-Cas включает транскрипцию CRISPR-локусов и сопутствующих генов Cas, кодирующих белки с нуклеазными и хеликазными доменами [42]. Эти белки используют crРНК для распознавания и инактивации чужеродных РНК и ДНК [40,43].
При повторном заражении crРНК объединяются с Cas-белками, формируя эффекторный комплекс [44]. Спейсерная последовательность в crРНК гибридизуется с комплементарной мишенью (например, вирусной ДНК или плазмидой), что позволяет комплексу распознать чужеродный генетический элемент [41,44]. Это взаимодействие запускает специфичное расщепление мишени Cas-нуклеазами [41], предотвращая распространение чужеродных нуклеиновых кислот.
3.1. Технология CRISPR-Cas9
Системы CRISPR-Cas делятся на шесть типов (I-VI), относящихся к двум классам, которые используют разные механизмы распознавания и расщепления мишеней [41]:
- Класс 1 (типы I, III, IV): многобелковые эффекторные комплексы crРНК
- Класс 2 (типы II, V, VI): одноцепочечные эффекторные комплексы crРНК
Типы I, II и V нацелены на ДНК, тогда как типы III и VI — на РНК [46]. В системах I и III для распознавания и расщепления мишени используется комплекс из нескольких Cas-белков [41,42]. В отличие от них, система типа II использует единственный белок — эндонуклеазу Cas9 — для специфичного расщепления ДНК [41].
Ключевым элементом является мотив PAM (protospacer-adjacent motif) — короткая последовательность, прилегающая к мишени и необходимая для ее распознавания большинством CRISPR-Cas систем, особенно типа II [41,42].
В системе CRISPR-Cas9 типа II:
1. Малая некодирующая tracrРНК связывается с повтором в crРНК [41]
2. Образуется дуплекс crРНК-tracrРНК, направляющий Cas9 к мишени
3. Cas9 расщепляет ДНК, содержащую комплементарный спейсеру участок и соседний PAM
Изменяя последовательность спейсера в guideРНК (гидРНК), можно запрограммировать систему на распознавание практически любого участка генома. Двухцепочечный разрыв затем репарируется:
- Случайно (с образованием инделов)
- Или направленно (с использованием матрицы для гомологичной рекомбинации)
Это позволяет проводить точные геномные модификации, включая вставку целевых генов (Рисунок 4).
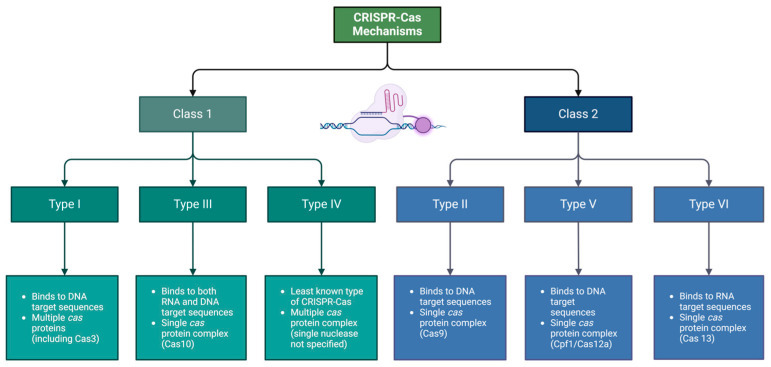
Дифференциация механизмов CRISPR-Cas
Механизмы CRISPR-Cas выполняют схожие функции, но различаются по структуре и/или принципу действия. Они подразделяются на два класса:
Класс 1: включает более сложные механизмы с мультисубъединичными эффекторными комплексами crРНК
Класс 2: состоит из менее сложных механизмов, использующих односубъединичные эффекторные комплексы crРНК
С начала 2000-х годов система CRISPR-Cas9 доказала свою эффективность как технология направленного редактирования генома на основе РНК, а также нашла применение в других областях, таких как целевой мутагенез, визуализация генома, эпигенетическая модуляция и регуляция транскрипции [41]. Проще говоря, технология CRISPR-Cas9 состоит из двух основных компонентов, которые могут доставляться в виде единой плазмиды при использовании в генной инженерии. К ним относятся: (a) бактериальный эндонуклеазный белок Cas9 и (b) специально разработанная однонаправляющая РНК (sgRNA), содержащая последовательность протоспейсера, гомологичную целевой ДНК. Кроме того, для лучшего понимания эффективности системы CRISPR-Cas9 и ее способности к связыванию необходимо углубить знания о влиянии доступности хроматина, а также эпигеномного окружения в целевом сайте [47].
3.2. Применение технологии CRISPR-Cas9 в ускоренной селекции
В данном разделе рассматривается использование технологии CRISPR-Cas9 для улучшения сельскохозяйственных культур и продуктов (например, повышение устойчивости к болезням и патогенам, увеличение срока хранения, устойчивость к потемнению [47]) с помощью генной инженерии и селекции растений. Генная инженерия в сочетании с методами ускоренной селекции может значительно ускорить процесс улучшения и выведения новых сортов.
У растений систему CRISPR-Cas9 можно доставлять различными методами, включая временные и трансформационные платформы, такие как трансфекция протопластов, агроинфильтрация и биобаллистическая доставка (бомбардировка частицами) в каллусные ткани. Многие исследовательские группы продемонстрировали, что технология CRISPR-Cas9 успешно применяется для трансформации модельных видов (например, Arabidopsis и табака), а также основных сельскохозяйственных культур, включая рис, пшеницу, сорго, кукурузу, томат, картофель и сою — все эти случаи были ранее обобщены [47]. Несмотря на успешность, процесс редактирования генов остается трудоемким, требующим культивирования тканей и специализированных лабораторий с физически изолированными зонами для доставки и работы системы CRISPR-Cas9 [26]. Однако инновационные системы, такие как ExpressEdit (впервые описанный Hickey et al. [26]), интегрируют быстрое редактирование генов непосредственно в программу ускоренной селекции, сокращая временные ограничения традиционного редактирования за счет исключения этапа in vitro. ExpressEdit пока не стал рутинной практикой, но он имеет огромное значение для будущего ускоренной селекции и улучшения сельскохозяйственных культур. Этот подход предполагает прямое внесение гена Cas9 и синтезированной sgRNA в растения. Например, такая sgRNA может придать признак устойчивости к болезням. Потомство обработанных растений можно проверить на наличие нового признака, и те растения, которые его проявляют и не содержат Cas9, могут быть отобраны [26]. Эти растения будут неотличимы от тех, которые получили признак в результате естественной мутации, и могут пройти традиционные этапы селекции, включая полевые испытания и выпуск сорта, избегая при этом маркировки как генетически модифицированных организмов. Альтернативно, Cas9 может оставаться в некоторых растениях, превращая их в «CRISPR-готовые» растения, которые могут быть использованы для дальнейших циклов редактирования генов путем внесения новых sgRNA, нацеленных на другие признаки [26].
Liu et al. недавно провели исследование, в котором использовали CRISPR-Cas9 для создания линий гибридных семян волосистого томата [48]. Ключом к успешному получению гибридных семян является создание мужско-стерильных линий, которые позволят сохранить чистоту линий томатов. Ранее этого можно было добиться только с помощью трудоемких процессов кастрации, однако внедрение системы CRISPR-Cas9 в этот процесс позволило проводить маркер-ассоциированный скрининг, инициирующий мутации, которые обеспечили мужскую стерильность у гибридных семян волосистого томата [48].
Как кратко обсуждалось выше, существует несколько способов доставки системы CRISPR-Cas9 в растения для направленного редактирования генома. Это также относится к подходам, не требующим культивирования тканей. Например, в качестве целевой ткани могут использоваться зародыши или протопласты. Однако, чтобы избежать этапа культивирования тканей, этот быстрый метод редактирования генов в идеале должен нацеливаться на зрелые семена или проростки [26,28]. В последующих поколениях фенотипирование также позволит осуществлять "накопление" (стэкинг) признаков [26,49].
Еще один метод включает использование наночастиц глины. Существует потенциал для инженерии глиняных нанопластин в качестве средства доставки как белка Cas9, так и, например, конструкции РНК-интерференции (RNAi) для устойчивости к вирусам [50]. Обычно эти компоненты доставляются с помощью вирусных векторов (например, геминивирусов) [51] или биобаллистическим методом (бомбардировка частицами) в апикальные меристемы побегов, что также позволяет редактировать пыльцу и ткани соцветий [26,49].
Механизмы CRISPR-Cas выполняют схожие функции, но различаются по структуре и/или принципу действия. Они подразделяются на два класса:
Класс 1: включает более сложные механизмы с мультисубъединичными эффекторными комплексами crРНК
Класс 2: состоит из менее сложных механизмов, использующих односубъединичные эффекторные комплексы crРНК
С начала 2000-х годов система CRISPR-Cas9 доказала свою эффективность как технология направленного редактирования генома на основе РНК, а также нашла применение в других областях, таких как целевой мутагенез, визуализация генома, эпигенетическая модуляция и регуляция транскрипции [41]. Проще говоря, технология CRISPR-Cas9 состоит из двух основных компонентов, которые могут доставляться в виде единой плазмиды при использовании в генной инженерии. К ним относятся: (a) бактериальный эндонуклеазный белок Cas9 и (b) специально разработанная однонаправляющая РНК (sgRNA), содержащая последовательность протоспейсера, гомологичную целевой ДНК. Кроме того, для лучшего понимания эффективности системы CRISPR-Cas9 и ее способности к связыванию необходимо углубить знания о влиянии доступности хроматина, а также эпигеномного окружения в целевом сайте [47].
3.2. Применение технологии CRISPR-Cas9 в ускоренной селекции
В данном разделе рассматривается использование технологии CRISPR-Cas9 для улучшения сельскохозяйственных культур и продуктов (например, повышение устойчивости к болезням и патогенам, увеличение срока хранения, устойчивость к потемнению [47]) с помощью генной инженерии и селекции растений. Генная инженерия в сочетании с методами ускоренной селекции может значительно ускорить процесс улучшения и выведения новых сортов.
У растений систему CRISPR-Cas9 можно доставлять различными методами, включая временные и трансформационные платформы, такие как трансфекция протопластов, агроинфильтрация и биобаллистическая доставка (бомбардировка частицами) в каллусные ткани. Многие исследовательские группы продемонстрировали, что технология CRISPR-Cas9 успешно применяется для трансформации модельных видов (например, Arabidopsis и табака), а также основных сельскохозяйственных культур, включая рис, пшеницу, сорго, кукурузу, томат, картофель и сою — все эти случаи были ранее обобщены [47]. Несмотря на успешность, процесс редактирования генов остается трудоемким, требующим культивирования тканей и специализированных лабораторий с физически изолированными зонами для доставки и работы системы CRISPR-Cas9 [26]. Однако инновационные системы, такие как ExpressEdit (впервые описанный Hickey et al. [26]), интегрируют быстрое редактирование генов непосредственно в программу ускоренной селекции, сокращая временные ограничения традиционного редактирования за счет исключения этапа in vitro. ExpressEdit пока не стал рутинной практикой, но он имеет огромное значение для будущего ускоренной селекции и улучшения сельскохозяйственных культур. Этот подход предполагает прямое внесение гена Cas9 и синтезированной sgRNA в растения. Например, такая sgRNA может придать признак устойчивости к болезням. Потомство обработанных растений можно проверить на наличие нового признака, и те растения, которые его проявляют и не содержат Cas9, могут быть отобраны [26]. Эти растения будут неотличимы от тех, которые получили признак в результате естественной мутации, и могут пройти традиционные этапы селекции, включая полевые испытания и выпуск сорта, избегая при этом маркировки как генетически модифицированных организмов. Альтернативно, Cas9 может оставаться в некоторых растениях, превращая их в «CRISPR-готовые» растения, которые могут быть использованы для дальнейших циклов редактирования генов путем внесения новых sgRNA, нацеленных на другие признаки [26].
Liu et al. недавно провели исследование, в котором использовали CRISPR-Cas9 для создания линий гибридных семян волосистого томата [48]. Ключом к успешному получению гибридных семян является создание мужско-стерильных линий, которые позволят сохранить чистоту линий томатов. Ранее этого можно было добиться только с помощью трудоемких процессов кастрации, однако внедрение системы CRISPR-Cas9 в этот процесс позволило проводить маркер-ассоциированный скрининг, инициирующий мутации, которые обеспечили мужскую стерильность у гибридных семян волосистого томата [48].
Как кратко обсуждалось выше, существует несколько способов доставки системы CRISPR-Cas9 в растения для направленного редактирования генома. Это также относится к подходам, не требующим культивирования тканей. Например, в качестве целевой ткани могут использоваться зародыши или протопласты. Однако, чтобы избежать этапа культивирования тканей, этот быстрый метод редактирования генов в идеале должен нацеливаться на зрелые семена или проростки [26,28]. В последующих поколениях фенотипирование также позволит осуществлять "накопление" (стэкинг) признаков [26,49].
Еще один метод включает использование наночастиц глины. Существует потенциал для инженерии глиняных нанопластин в качестве средства доставки как белка Cas9, так и, например, конструкции РНК-интерференции (RNAi) для устойчивости к вирусам [50]. Обычно эти компоненты доставляются с помощью вирусных векторов (например, геминивирусов) [51] или биобаллистическим методом (бомбардировка частицами) в апикальные меристемы побегов, что также позволяет редактировать пыльцу и ткани соцветий [26,49].
Технология CRISPR-Cas также была применена для индукции синтетического апомиксиса у растений с целью улучшения гибридной силы в программах селекции сельскохозяйственных культур [52]. Апомиксис — это соматический метод бесполого размножения, при котором образуется клон, генетически идентичный материнскому растению [53]. В последнее время индукция синтетического апомиксиса с помощью колхицин-опосредованного мутагенеза изучалась на примере египетского гибрида риса, что привело к фиксации гетерозиса, который обычно нарушается из-за расщепления в популяции [53]. Однако одной из наиболее актуальных целей развития признаков апомиксиса в селекционных линиях растений является обеспечение трансгенерационной экспрессии фенотипов, полученных с помощью трансгенных или традиционных методов селекции. Это продемонстрировали Liu et al. [54], которым удалось добиться трансгенерационной экспрессии фенотипов у гибрида риса путем индукции синтетического апомиксиса.
Традиционные методы трансформации сельскохозяйственных растений, уже обладающих признаками апомиксиса, часто демонстрируют различия в ближайших поколениях, но требуют множества испытаний, прежде чем трансгены станут заметными и функциональными в последующих поколениях, как показали Aragão et al. [55]. Однако было обнаружено, что эта тенденция меняется при использовании более инновационных систем трансформации. Например, Feng et al. [56] исследовали наследуемость CRISPR-Cas-индуцированных генетических изменений и обнаружили, что, хотя некоторые модификации не наследуются у исходных растений, они могут проявляться в последующих поколениях.
В настоящее время для улучшения сельскохозяйственных культур успешно клонировано несколько генов, многие из которых являются генами устойчивости (R-генами) к различным биотическим и абиотическим стрессам [57]. Ген Helminthosporium carbonum susceptibility 1 (Hm1) у кукурузы, кодирующий детоксикацию грибкового патогена Cochliobolus carbonum, стал первым клонированным R-геном [58] и открыл путь для клонирования R-генов в селекции растений. Аналогично, Luo et al. [59] провели новаторское исследование, которое привело к идентификации стэка из пяти трансгенов, способного обеспечивать генетически связанную широкоспектральную устойчивость к патогену стеблевой ржавчины пшеницы Puccinia graminis f. sp. Tritici. В исследованиях R-генов абиотического стресса у риса (Oryza sativa L.) продвинутое QTL-картирование позволило выявить несколько генов, связанных с солеустойчивостью, что заложило основу для их клонирования и молекулярно-ассистированной селекции [60]. Улучшение иммунитета растений за счет внедрения генов устойчивости помогает ускорить селекционный процесс, защищая культуры от разрушительных заболеваний и неблагоприятных условий, тем самым сохраняя потенциальную урожайность.
Редактирование генов с помощью технологии CRISPR-Cas9 в сочетании с ускоренной селекцией предоставляет мощный инструмент для создания сортов сельскохозяйственных культур следующего поколения, способных к высокой урожайности и устойчивости к стрессам. Такие культуры имеют большой потенциал для удовлетворения растущих потребностей населения.
4. Геномный отбор
С постоянным развитием полевых и лабораторных методик, таких как применение светодиодов и CRISPR/Cas9, необходимо также учитывать потенциал компьютерных (in silico) схем ускоренной селекции. Значительные достижения в области высокоплотного секвенирования генома и вычислительных биоинформационных технологий предоставили селекционерам растений мощные инструменты для быстрого выведения новых сортов. Геномный отбор (GS), также называемый полногеномным отбором или геномным прогнозированием, представляет собой один из таких инструментов.
4.1. Обзор методологии геномного отбора
Цель геномного отбора - выявление желаемых признаков у особей или линий целевого вида без необходимости длительных полевых испытаний или фенотипирования. Этот процесс проводится в две основные фазы:
1. Фаза обучения:
- Отбирается популяция особей на основе их генетической близости к селекционным популяциям, а также их способности представлять потенциальное генетическое разнообразие этой популяции.
- Эти особи генотипируются с использованием сотен тысяч полногеномных маркеров и фенотипируются по интересующему признаку или признакам [41].
- Собранные данные используются для широкой корреляции генотипа с фенотипом с помощью созданной прогностической модели.
- Две подгруппы популяций составляют эту фазу: обучающая популяция, на основе которой формируется модель, и валидационная популяция, используемая для проверки точности модели [51].
2. Фаза отбора:
- Эта модель затем используется во второй фазе: отборе из селекционной популяции.
- На этом этапе модель GS может предоставить геномно-оцененные селекционные значения (GEBVs) на основе ввода геномных данных и прогнозируемых эффектов маркеров.
- Линии селекционной популяции требуют только генотипирования для отбора особей для продвижения или исключения из селекционной программы [61,62].
4.2. Метод геномного отбора для программ ускоренной селекции
Геномный отбор, впервые предложенный Meuwissen et al. [63], получил широкое распространение в программах разведения молочного скота в США, поскольку этот метод позволяет селекционерам оценивать племенную ценность производителей без необходимости ждать годы для созревания и фенотипирования потомства. Потенциальные преимущества этого метода разведения для полевых культур, таких как кукуруза, пшеница и рис, были оценены в исследованиях по селекции растений в ограниченной степени и еще не используются в полной мере [62]. Поверхностное сходство GS с часто используемыми методами, такими как маркер-ассоциированный отбор (MAS), а также первоначальные затраты способствовали медленному внедрению этого метода селекционерами растений. Однако преимущества этой методики в программе ускоренной селекции необходимо учитывать.
Обычные методы молекулярной селекции, такие как MAS, требуют детального знания биологической функции признака и генетики признака, что приводит к подходу "селекция по проекту", требующему многолетних исследований и валидации перед внедрением. Использование полногеномных прогностических моделей в геномном отборе устраняет это ограничение, одновременно учитывая группы предикторов, а не отдельные локусы количественных признаков (QTL) [64]. Это позволяет селекционным программам отбирать признаки, которые еще не изучены подробно. Кроме того, MAS ограничен в своей способности идентифицировать сложные, полигенные признаки. Это оставляет многие ключевые хозяйственно-ценные признаки, такие как урожайность и устойчивость, недостижимыми только с помощью MAS. Модели геномного отбора, с другой стороны, способны улавливать большинство локусов с малым эффектом, которые вносят вклад в целевой признак, при условии, что локусы признаков и полногеномные маркеры находятся в некоторой степени неравновесного сцепления [65].
Как упоминалось ранее, одним из наиболее заметных преимуществ геномного отбора является значительное сокращение времени, необходимого для получения больших генетических достижений. Полевые испытания для точного фенотипирования селекционных популяций требуют многолетних дорогостоящих испытаний в различных условиях и местах; эта проблема преодолевается геномным отбором благодаря способности модели предсказывать желаемые признаки просто на основе наблюдения за генотипом [66]. Ярким примером является масличная пальма (Elais guineensis), где было продемонстрировано сокращение цикла отбора в селекционной программе с 19 до 6 лет после внедрения GS [64]. Время и средства, необходимые для полевых испытаний селекционных популяций, могут быть значительно сокращены, что приведет к большей генетической прибыли за единицу времени. По мере того как базы данных последовательностей культур продолжают расширяться, ресурсы, доступные селекционерам, позволяют внедрять геномный отбор с большей точностью и гибкостью в проектировании статистических моделей.
Хотя методы селекции с использованием GS обладают огромным потенциалом для ускорения селекционного процесса, эффективность геномного отбора и прогнозирования в селекции растений может ограничиваться сложностью целевых генов и полигенных комплексов признаков [67]. Полная эффективность этого селекционного инструмента требует учета всех потенциальных ограничений, включая наследуемость и сложную генетическую архитектуру. Однако в решении этой проблемы уже достигнут значительный прогресс. Недавно время приготовления различных популяций фасоли обыкновенной (Phaseolus vulgaris L.) - сложный признак, затрагивающий несколько геномных регионов, - было успешно предсказано с помощью методов GS-анализа [68]. Учитывая сложность признака времени приготовления, применение методов GS позволило провести оперативный анализ задействованных геномных путей. Хотя применение GS в программах ускоренной селекции пока ограничено из-за недостатка прецедентов в сообществе селекционеров растений и высоких начальных затрат на секвенирование, эти барьеры будут постепенно преодолеваться по мере того, как больше исследователей начнут изучать потенциал этой методики.
5. Эпигеномика/Эпигенетика
Эпигеномика определяется как совокупность всех изменений в экспрессии генов и клеточных функциях, вызванных модификациями ДНК и гистонов, а также синтезом некодирующих РНК без изменения основной последовательности ДНК [69]. Модификации ДНК и гистонов играют ключевую роль в регуляции экспрессии генов и, соответственно, в развитии растений и их реакции на окружающую среду [70]. Наиболее распространенные примеры таких модификаций включают метилирование ДНК, ацетилирование, убиквитинирование и фосфорилирование гистонов. Метилирование ДНК является одним из наиболее изученных эпигенетических маркеров у растений [71]. Этот тип модификации, как и другие, может быть отслежен и картирован с помощью технологий высокопроизводительного секвенирования нового поколения, что позволяет глубже понять роль модификаций гистонов и ДНК в регуляции роста и развития растений, а также их реакций на биотические и абиотические стрессы [72]. Метилирование цитозина происходит по пятому положению цитозинового основания с помощью фермента ДНК-метилтрансферазы и проявляется в трех различных контекстах нуклеотидных последовательностей: CG, CHG и CHH [73]. Эти паттерны наследуются при клеточном делении и могут влиять на фенотипическую изменчивость.
Паттерны метилирования ДНК могут изменяться под воздействием факторов окружающей среды, таких как абиотические и биотические стрессоры, или вследствие генетических факторов [74]. Расшифровка паттернов метилирования ДНК и генетической информации об эпигенетических маркерах может дать представление о том, как эпиаллели контролируют экспрессию генов и транскриптов на разных стадиях развития растения и его реакции на стресс у различных генотипов сельскохозяйственных культур [72]. Паттерн CG, также известный как генное метилирование (gbM), относится к метилированию, происходящему в транскрипционных регионах и связанному с умеренными уровнями экспрессии. Паттерн CG-подобного метилирования транспозонных элементов (TE) ассоциирован с подавлением активности TE (Рисунок 5). Активность транспозонных элементов играет важную роль в увеличении геномного разнообразия, что делает их перспективными для селекции [33].
Традиционные методы трансформации сельскохозяйственных растений, уже обладающих признаками апомиксиса, часто демонстрируют различия в ближайших поколениях, но требуют множества испытаний, прежде чем трансгены станут заметными и функциональными в последующих поколениях, как показали Aragão et al. [55]. Однако было обнаружено, что эта тенденция меняется при использовании более инновационных систем трансформации. Например, Feng et al. [56] исследовали наследуемость CRISPR-Cas-индуцированных генетических изменений и обнаружили, что, хотя некоторые модификации не наследуются у исходных растений, они могут проявляться в последующих поколениях.
В настоящее время для улучшения сельскохозяйственных культур успешно клонировано несколько генов, многие из которых являются генами устойчивости (R-генами) к различным биотическим и абиотическим стрессам [57]. Ген Helminthosporium carbonum susceptibility 1 (Hm1) у кукурузы, кодирующий детоксикацию грибкового патогена Cochliobolus carbonum, стал первым клонированным R-геном [58] и открыл путь для клонирования R-генов в селекции растений. Аналогично, Luo et al. [59] провели новаторское исследование, которое привело к идентификации стэка из пяти трансгенов, способного обеспечивать генетически связанную широкоспектральную устойчивость к патогену стеблевой ржавчины пшеницы Puccinia graminis f. sp. Tritici. В исследованиях R-генов абиотического стресса у риса (Oryza sativa L.) продвинутое QTL-картирование позволило выявить несколько генов, связанных с солеустойчивостью, что заложило основу для их клонирования и молекулярно-ассистированной селекции [60]. Улучшение иммунитета растений за счет внедрения генов устойчивости помогает ускорить селекционный процесс, защищая культуры от разрушительных заболеваний и неблагоприятных условий, тем самым сохраняя потенциальную урожайность.
Редактирование генов с помощью технологии CRISPR-Cas9 в сочетании с ускоренной селекцией предоставляет мощный инструмент для создания сортов сельскохозяйственных культур следующего поколения, способных к высокой урожайности и устойчивости к стрессам. Такие культуры имеют большой потенциал для удовлетворения растущих потребностей населения.
4. Геномный отбор
С постоянным развитием полевых и лабораторных методик, таких как применение светодиодов и CRISPR/Cas9, необходимо также учитывать потенциал компьютерных (in silico) схем ускоренной селекции. Значительные достижения в области высокоплотного секвенирования генома и вычислительных биоинформационных технологий предоставили селекционерам растений мощные инструменты для быстрого выведения новых сортов. Геномный отбор (GS), также называемый полногеномным отбором или геномным прогнозированием, представляет собой один из таких инструментов.
4.1. Обзор методологии геномного отбора
Цель геномного отбора - выявление желаемых признаков у особей или линий целевого вида без необходимости длительных полевых испытаний или фенотипирования. Этот процесс проводится в две основные фазы:
1. Фаза обучения:
- Отбирается популяция особей на основе их генетической близости к селекционным популяциям, а также их способности представлять потенциальное генетическое разнообразие этой популяции.
- Эти особи генотипируются с использованием сотен тысяч полногеномных маркеров и фенотипируются по интересующему признаку или признакам [41].
- Собранные данные используются для широкой корреляции генотипа с фенотипом с помощью созданной прогностической модели.
- Две подгруппы популяций составляют эту фазу: обучающая популяция, на основе которой формируется модель, и валидационная популяция, используемая для проверки точности модели [51].
2. Фаза отбора:
- Эта модель затем используется во второй фазе: отборе из селекционной популяции.
- На этом этапе модель GS может предоставить геномно-оцененные селекционные значения (GEBVs) на основе ввода геномных данных и прогнозируемых эффектов маркеров.
- Линии селекционной популяции требуют только генотипирования для отбора особей для продвижения или исключения из селекционной программы [61,62].
4.2. Метод геномного отбора для программ ускоренной селекции
Геномный отбор, впервые предложенный Meuwissen et al. [63], получил широкое распространение в программах разведения молочного скота в США, поскольку этот метод позволяет селекционерам оценивать племенную ценность производителей без необходимости ждать годы для созревания и фенотипирования потомства. Потенциальные преимущества этого метода разведения для полевых культур, таких как кукуруза, пшеница и рис, были оценены в исследованиях по селекции растений в ограниченной степени и еще не используются в полной мере [62]. Поверхностное сходство GS с часто используемыми методами, такими как маркер-ассоциированный отбор (MAS), а также первоначальные затраты способствовали медленному внедрению этого метода селекционерами растений. Однако преимущества этой методики в программе ускоренной селекции необходимо учитывать.
Обычные методы молекулярной селекции, такие как MAS, требуют детального знания биологической функции признака и генетики признака, что приводит к подходу "селекция по проекту", требующему многолетних исследований и валидации перед внедрением. Использование полногеномных прогностических моделей в геномном отборе устраняет это ограничение, одновременно учитывая группы предикторов, а не отдельные локусы количественных признаков (QTL) [64]. Это позволяет селекционным программам отбирать признаки, которые еще не изучены подробно. Кроме того, MAS ограничен в своей способности идентифицировать сложные, полигенные признаки. Это оставляет многие ключевые хозяйственно-ценные признаки, такие как урожайность и устойчивость, недостижимыми только с помощью MAS. Модели геномного отбора, с другой стороны, способны улавливать большинство локусов с малым эффектом, которые вносят вклад в целевой признак, при условии, что локусы признаков и полногеномные маркеры находятся в некоторой степени неравновесного сцепления [65].
Как упоминалось ранее, одним из наиболее заметных преимуществ геномного отбора является значительное сокращение времени, необходимого для получения больших генетических достижений. Полевые испытания для точного фенотипирования селекционных популяций требуют многолетних дорогостоящих испытаний в различных условиях и местах; эта проблема преодолевается геномным отбором благодаря способности модели предсказывать желаемые признаки просто на основе наблюдения за генотипом [66]. Ярким примером является масличная пальма (Elais guineensis), где было продемонстрировано сокращение цикла отбора в селекционной программе с 19 до 6 лет после внедрения GS [64]. Время и средства, необходимые для полевых испытаний селекционных популяций, могут быть значительно сокращены, что приведет к большей генетической прибыли за единицу времени. По мере того как базы данных последовательностей культур продолжают расширяться, ресурсы, доступные селекционерам, позволяют внедрять геномный отбор с большей точностью и гибкостью в проектировании статистических моделей.
Хотя методы селекции с использованием GS обладают огромным потенциалом для ускорения селекционного процесса, эффективность геномного отбора и прогнозирования в селекции растений может ограничиваться сложностью целевых генов и полигенных комплексов признаков [67]. Полная эффективность этого селекционного инструмента требует учета всех потенциальных ограничений, включая наследуемость и сложную генетическую архитектуру. Однако в решении этой проблемы уже достигнут значительный прогресс. Недавно время приготовления различных популяций фасоли обыкновенной (Phaseolus vulgaris L.) - сложный признак, затрагивающий несколько геномных регионов, - было успешно предсказано с помощью методов GS-анализа [68]. Учитывая сложность признака времени приготовления, применение методов GS позволило провести оперативный анализ задействованных геномных путей. Хотя применение GS в программах ускоренной селекции пока ограничено из-за недостатка прецедентов в сообществе селекционеров растений и высоких начальных затрат на секвенирование, эти барьеры будут постепенно преодолеваться по мере того, как больше исследователей начнут изучать потенциал этой методики.
5. Эпигеномика/Эпигенетика
Эпигеномика определяется как совокупность всех изменений в экспрессии генов и клеточных функциях, вызванных модификациями ДНК и гистонов, а также синтезом некодирующих РНК без изменения основной последовательности ДНК [69]. Модификации ДНК и гистонов играют ключевую роль в регуляции экспрессии генов и, соответственно, в развитии растений и их реакции на окружающую среду [70]. Наиболее распространенные примеры таких модификаций включают метилирование ДНК, ацетилирование, убиквитинирование и фосфорилирование гистонов. Метилирование ДНК является одним из наиболее изученных эпигенетических маркеров у растений [71]. Этот тип модификации, как и другие, может быть отслежен и картирован с помощью технологий высокопроизводительного секвенирования нового поколения, что позволяет глубже понять роль модификаций гистонов и ДНК в регуляции роста и развития растений, а также их реакций на биотические и абиотические стрессы [72]. Метилирование цитозина происходит по пятому положению цитозинового основания с помощью фермента ДНК-метилтрансферазы и проявляется в трех различных контекстах нуклеотидных последовательностей: CG, CHG и CHH [73]. Эти паттерны наследуются при клеточном делении и могут влиять на фенотипическую изменчивость.
Паттерны метилирования ДНК могут изменяться под воздействием факторов окружающей среды, таких как абиотические и биотические стрессоры, или вследствие генетических факторов [74]. Расшифровка паттернов метилирования ДНК и генетической информации об эпигенетических маркерах может дать представление о том, как эпиаллели контролируют экспрессию генов и транскриптов на разных стадиях развития растения и его реакции на стресс у различных генотипов сельскохозяйственных культур [72]. Паттерн CG, также известный как генное метилирование (gbM), относится к метилированию, происходящему в транскрипционных регионах и связанному с умеренными уровнями экспрессии. Паттерн CG-подобного метилирования транспозонных элементов (TE) ассоциирован с подавлением активности TE (Рисунок 5). Активность транспозонных элементов играет важную роль в увеличении геномного разнообразия, что делает их перспективными для селекции [33].
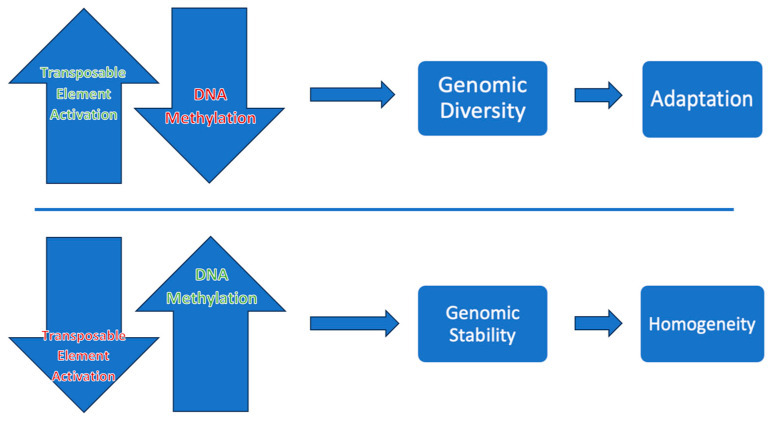
Эпигенетическая взаимосвязь между активностью транспозонных элементов (TE) и метилированием ДНК. Когда уровень метилирования ДНК ниже, активация TE выше, что способствует геномному разнообразию. Подавление активации TE способствует геномной стабильности.
Взаимосвязь между метилированием ДНК и экспрессией генов может быть использована путем целенаправленного нарушения метилирования ДНК с помощью внешних воздействий, что может помочь ускорить создание изменчивости для улучшения сельскохозяйственных культур. Этого можно достичь с помощью методов, аналогичных тем, которые используются в мутационной селекции, или путем применения ингибиторов ДНК-метилтрансфераз, что приводит к геномному деметилированию ДНК. Также могут быть использованы методы направленного воздействия на эпигеном, способные изменять метилирование ДНК, что может привести к фенотипическим изменениям, а также к изменениям в экспрессии генов [75].
Взаимосвязь между метилированием ДНК и экспрессией генов может быть использована путем целенаправленного нарушения метилирования ДНК с помощью внешних воздействий, что может помочь ускорить создание изменчивости для улучшения сельскохозяйственных культур. Этого можно достичь с помощью методов, аналогичных тем, которые используются в мутационной селекции, или путем применения ингибиторов ДНК-метилтрансфераз, что приводит к геномному деметилированию ДНК. Также могут быть использованы методы направленного воздействия на эпигеном, способные изменять метилирование ДНК, что может привести к фенотипическим изменениям, а также к изменениям в экспрессии генов [75].
Известным методом эпигенетической инженерии является РНК-интерференция (RNAi). RNAi представляет собой метод посттранскрипционного сайленсинга генов, осуществляемый с помощью экспрессии двуцепочечной РНК (дцРНК) [76]. Малые интерферирующие РНК используются для усиления экспрессии конкретного гена в его транскриптоме, способствуя проявлению определенного признака [77]. Расщепление дцРНК (например, микроРНК и малых интерферирующих РНК) на активные малые некодирующие РНК осуществляется с помощью мультидоменной эндорибонуклеазы Dicer (или Dicer-подобных белков) [78]. Эти малые некодирующие РНК, участвующие в процессе регуляции генов, включаются во вторую рибонуклеазную систему, состоящую из РНК-индуцированного комплекса сайленсинга (RISC), а также аргонавт-белков и других эффекторных белков, которая инициирует процесс сайленсинга генов путем направленной деградации матричных РНК [78,79]. Системы RNAi могут быть доставлены временно с помощью классических методов трансформации, включая биобаллистическую доставку (бомбардировку частицами), вирусные векторы и агробактериальную трансформацию [79].
Применение эпигенетики в селекции
Эпигенетическая инженерия может приводить к модификации признаков у растений без изменения последовательности ДНК. Было обнаружено несколько эпиаллелей, вызывающих фенотипическую и морфологическую изменчивость; однако этот процесс может занять некоторое время, поскольку зависит от спонтанных или естественных эпимутаций. Известно, что эпигенетические вариации влияют на важные признаки у сельскохозяйственных культур; поэтому «манипулирование стабильно наследуемой эпигенетической изменчивостью может стать мощным инструментом в селекции растений» [69]. Направленные ферменты, модифицирующие ДНК/хроматин, могут быть использованы для эпигенетического воздействия на метки, влияющие на экспрессию генов. Эти модифицирующие ферменты успешно изменяют эпигенетические метки в целевых участках. Они также используются для повышения и понижения транскрипции генов [69].
Технология RNAi применялась для решения различных типов и степеней биотических и абиотических стрессов [80]. Активируя аномальную экспрессию защитных генов против патогенов растений и других вредных факторов, RNAi способствовала созданию признаков, включая устойчивость к бактериям [81], грибам [82], насекомым [83], засухоустойчивость [84,85], термоустойчивость [86] и солеустойчивость [87]. RNAi позволяет оценивать функцию генов, создавать культуры, устойчивые к абиотическим и биотическим стрессам, разрабатывать устойчивые к болезням сорта [76,77,88], а также проводить метаболическую инженерию растений [78].
Понимание того, как эпигенетические изменения влияют на множественные копии генов, может дать представление о создании культур, устойчивых к множественным стрессам. Чтобы обеспечить способность растений выдерживать изменения окружающей среды, необходимо стабильное наследование адаптивных эпиаллелей. Как только наследование метилирования ДНК стабилизируется, эпиаллель также станет стабильным, что снизит образование новых эпиаллелей. Например, Ketumile et al. [89] обнаружили, что эпигенетическая изменчивость у сорго (Sorghum bicolor) продемонстрировала возможности селекции сорго на устойчивость за счет фокусировки на гене Mutator S HOMOLOG 1 (MSH1). В этом исследовании сайленсинг гена MSH1, индуцированный RNAi, вызвал межпоколенческие эпигеномные изменения — феномен, известный как репаттернинг метилирования [80,89,90]. Если модификация ДНК нестабильна, это может увеличить образование или потерю эпиаллелей в пределах одного поколения. Таким образом, стабильность эпигенетической метки в поколениях играет ключевую роль в ее способности способствовать улучшению сельскохозяйственных культур [69]. Два отдельных исследования показали, что Arabidopsis, подвергавшийся воздействию засоления [91], и рис, подвергавшийся засухе [92] в течение нескольких поколений, демонстрировали повышенные скорости изменений в метилировании ДНК, что указывает на то, что стабильность метилирования может изменяться под воздействием условий окружающей среды.
Селекция растений с помощью эпигенетической инженерии может занять несколько поколений для проявления фенотипической изменчивости или наследования желаемых признаков; поэтому использование ускоренной селекции в сочетании с эпигенетическими инструментами может сократить время генерации, одновременно создавая культуры, устойчивые к различным стрессам. Поскольку эпигенетическая регуляция экспрессии генов влияет на признаки сельскохозяйственных культур, она может быть использована в качестве мощного инструмента в селекции растений. Важным преимуществом является то, что она не затрагивает изменения последовательности ДНК и представляет собой естественный путь.
6. Обсуждение и перспективы развития
Для развития программ ускоренной селекции современные технологии всегда будут оставаться крайне востребованными. Внедрение технологий ускоренной селекции, подобных рассмотренным выше, уже стало необходимостью в мире с постоянно растущим населением и сокращающимися ресурсами. Методы и технологии ускоренной селекции предлагают решения проблем, которые годами сдерживали эффективный рост сельскохозяйственных культур.
Рассмотренные методы и примеры (Рисунок 6) не только устраняют факторы, замедляющие процессы селекции и производства сельскохозяйственных культур, но и корректируют аспекты растениеводческих систем, снижающие общую эффективность. Ускоренная селекция использует регулируемые условия окружающей среды, включая температуру, интенсивность освещения и контроль продолжительности светового дня, что приводит к сокращению времени генерации [93].
Внедрение технологий ускоренной селекции в коммерческие системы растениеводства позволит селекционерам смягчать последствия нехватки продовольствия, вызванной изменениями природных явлений, что представляет собой одно из наиболее негативных социальных последствий климатических изменений. Эти инновационные подходы открывают новые возможности для устойчивого сельского хозяйства, способного адаптироваться к глобальным вызовам современности, обеспечивая продовольственную безопасность в условиях меняющейся среды и растущих потребностей человечества.
Применение эпигенетики в селекции
Эпигенетическая инженерия может приводить к модификации признаков у растений без изменения последовательности ДНК. Было обнаружено несколько эпиаллелей, вызывающих фенотипическую и морфологическую изменчивость; однако этот процесс может занять некоторое время, поскольку зависит от спонтанных или естественных эпимутаций. Известно, что эпигенетические вариации влияют на важные признаки у сельскохозяйственных культур; поэтому «манипулирование стабильно наследуемой эпигенетической изменчивостью может стать мощным инструментом в селекции растений» [69]. Направленные ферменты, модифицирующие ДНК/хроматин, могут быть использованы для эпигенетического воздействия на метки, влияющие на экспрессию генов. Эти модифицирующие ферменты успешно изменяют эпигенетические метки в целевых участках. Они также используются для повышения и понижения транскрипции генов [69].
Технология RNAi применялась для решения различных типов и степеней биотических и абиотических стрессов [80]. Активируя аномальную экспрессию защитных генов против патогенов растений и других вредных факторов, RNAi способствовала созданию признаков, включая устойчивость к бактериям [81], грибам [82], насекомым [83], засухоустойчивость [84,85], термоустойчивость [86] и солеустойчивость [87]. RNAi позволяет оценивать функцию генов, создавать культуры, устойчивые к абиотическим и биотическим стрессам, разрабатывать устойчивые к болезням сорта [76,77,88], а также проводить метаболическую инженерию растений [78].
Понимание того, как эпигенетические изменения влияют на множественные копии генов, может дать представление о создании культур, устойчивых к множественным стрессам. Чтобы обеспечить способность растений выдерживать изменения окружающей среды, необходимо стабильное наследование адаптивных эпиаллелей. Как только наследование метилирования ДНК стабилизируется, эпиаллель также станет стабильным, что снизит образование новых эпиаллелей. Например, Ketumile et al. [89] обнаружили, что эпигенетическая изменчивость у сорго (Sorghum bicolor) продемонстрировала возможности селекции сорго на устойчивость за счет фокусировки на гене Mutator S HOMOLOG 1 (MSH1). В этом исследовании сайленсинг гена MSH1, индуцированный RNAi, вызвал межпоколенческие эпигеномные изменения — феномен, известный как репаттернинг метилирования [80,89,90]. Если модификация ДНК нестабильна, это может увеличить образование или потерю эпиаллелей в пределах одного поколения. Таким образом, стабильность эпигенетической метки в поколениях играет ключевую роль в ее способности способствовать улучшению сельскохозяйственных культур [69]. Два отдельных исследования показали, что Arabidopsis, подвергавшийся воздействию засоления [91], и рис, подвергавшийся засухе [92] в течение нескольких поколений, демонстрировали повышенные скорости изменений в метилировании ДНК, что указывает на то, что стабильность метилирования может изменяться под воздействием условий окружающей среды.
Селекция растений с помощью эпигенетической инженерии может занять несколько поколений для проявления фенотипической изменчивости или наследования желаемых признаков; поэтому использование ускоренной селекции в сочетании с эпигенетическими инструментами может сократить время генерации, одновременно создавая культуры, устойчивые к различным стрессам. Поскольку эпигенетическая регуляция экспрессии генов влияет на признаки сельскохозяйственных культур, она может быть использована в качестве мощного инструмента в селекции растений. Важным преимуществом является то, что она не затрагивает изменения последовательности ДНК и представляет собой естественный путь.
6. Обсуждение и перспективы развития
Для развития программ ускоренной селекции современные технологии всегда будут оставаться крайне востребованными. Внедрение технологий ускоренной селекции, подобных рассмотренным выше, уже стало необходимостью в мире с постоянно растущим населением и сокращающимися ресурсами. Методы и технологии ускоренной селекции предлагают решения проблем, которые годами сдерживали эффективный рост сельскохозяйственных культур.
Рассмотренные методы и примеры (Рисунок 6) не только устраняют факторы, замедляющие процессы селекции и производства сельскохозяйственных культур, но и корректируют аспекты растениеводческих систем, снижающие общую эффективность. Ускоренная селекция использует регулируемые условия окружающей среды, включая температуру, интенсивность освещения и контроль продолжительности светового дня, что приводит к сокращению времени генерации [93].
Внедрение технологий ускоренной селекции в коммерческие системы растениеводства позволит селекционерам смягчать последствия нехватки продовольствия, вызванной изменениями природных явлений, что представляет собой одно из наиболее негативных социальных последствий климатических изменений. Эти инновационные подходы открывают новые возможности для устойчивого сельского хозяйства, способного адаптироваться к глобальным вызовам современности, обеспечивая продовольственную безопасность в условиях меняющейся среды и растущих потребностей человечества.

Методы ускоренной селекции и инновации обладают потенциалом для значительного прогресса в отрасли селекции сельскохозяйственных культур, однако развитие этих технологий сопряжено с естественными трудностями и проблемами. Например, взаимодействие генотипа со средой в испытаниях геномной селекции осложняет селекционные усилия, снижая эффективность отбора лучших особей для разведения, поскольку эти взаимодействия влияют на стабильность урожайности в поколениях. Хотя влияние среды играет ключевую роль в этих взаимодействиях, сам процесс отбора в большей степени зависит от взаимосвязи между производственными условиями и условиями селекции [94]. Классические методы отбора обычно проводятся в идеальных, стерильных производственных условиях, что часто приводит к недостаточному учету возможного влияния менее предсказуемых аспектов изменчивости среды (например, климатических условий) на результаты селекционных испытаний. Взаимодействие генотипа со средой важно для выявления генетической основы, благодаря которой растения адаптируются к новым условиям [95]. Акцент на взаимодействие генотипа и среды у сельскохозяйственных культур позволяет и стимулирует селекционеров выбирать растения не только на основе прогнозируемой гетерозисной мощности, но и с учетом их универсальности или способности занимать определенную экологическую нишу в экосистеме [96].
Доступность и простота использования представляют дополнительные сложности для внедрения инноваций в ускоренную селекцию. Например, хотя системы геномной селекции (GS) в целом упрощают селекционный процесс, существует потребность в более стандартизированных программах, включая подготовку кадров для создания и использования моделей геномной селекции, методы сбора расширенных данных и ведение записей [97]. Отсутствие четких руководств затрудняет внедрение методов геномной селекции в исследования [98]. В то же время CRISPR-Cas9, благодаря своей популярности как удобного инструмента редактирования генома, становится все более доступным как в академической среде, так и в частном секторе [52,99]. Аналогично, технология РНК-интерференции (RNAi) доказала свою высокую применимость в ускоренной селекции и уже используется в сельском хозяйстве [100].
Разработка и совершенствование новых технологий, способствующих развитию ускоренной селекции, может проводиться на различных видах растений, однако зачастую проще разработать стандарт, тестируя новые методы на хорошо изученных видах. Именно поэтому кукуруза (*Zea mays L.*) часто используется в качестве модельного растения для испытания новых агротехнологий. Например, Сингх и др. [101] использовали кукурузу для моделирования условий освещения и температуры при ускоренной селекции, обнаружив, что она способна выдерживать более высокую интенсивность света по сравнению с другими растениями, что позволяет ей адаптироваться к уровню освещенности, характерному для условий ускоренной селекции. Кукуруза также обладает высокой эффективностью использования радиации, определяемой как количество микромолей биомассы, произведенной на моль фотосинтетически активных фотонов, поглощенных зелеными частями растения [101]. Кроме того, урожайность кукурузы за сезон сильно зависит от идеальных климатических условий, которые нарушаются под воздействием абиотических стрессов [102]. Кукуруза была использована в качестве объекта 16-линейного мультиродительского скрещивания (MAGIC) для дальнейшего изучения генетической основы взаимодействия генотипа со средой. Шестнадцать линий кукурузы исследовались в пяти умеренных зонах с контрастными климатическими условиями и различными методами управления [95]. Общие характеристики роста кукурузы, ее чувствительность к факторам среды и история использования в качестве модели в исследованиях растений [103,104] укрепили ее позиции в качестве основной культуры для тестирования и разработки технологий ускоренной селекции.
Будущие исследования могут стать пионерами в интеграции ключевых молекулярных методов, таких как геномная селекция, CRISPR-Cas9 и эпигенетическая инженерия, с растущими массивами данных из различных направлений «омиксных» исследований — от транскриптомики до протеомики — для создания системы комплексных селекционных подходов. В перспективе разработка технологии генотипирования MiMe (митоз вместо мейоза) демонстрирует потенциал для успешного внедрения в системы ускоренной селекции [52]. Дополнительные исследования апомиксиса у мутантных форм сельскохозяйственных культур также открывают путь для изучения наследования признаков в поколениях, что повышает эффективность клонального размножения растений [52]. В сочетании с оптимизированными условиями выращивания, сокращающими время генерации, эти технологии будут непрерывно улучшать способность селекционеров быстро создавать высококачественные и устойчивые сорта, способные отвечать вызовам растущего мира.
Вклад авторов
M.S. conceived the work; L.W.L., M.P. and K.W. performed the searches and wrote the manuscript, with contributions from M.S., A.T. and G.O. All authors have read and agreed to the published version of the manuscript.
Конфликт интересов
The authors declare no conflicts of interest.
Финансирование
This research received no external funding.
Примечания
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Литература
Доступность и простота использования представляют дополнительные сложности для внедрения инноваций в ускоренную селекцию. Например, хотя системы геномной селекции (GS) в целом упрощают селекционный процесс, существует потребность в более стандартизированных программах, включая подготовку кадров для создания и использования моделей геномной селекции, методы сбора расширенных данных и ведение записей [97]. Отсутствие четких руководств затрудняет внедрение методов геномной селекции в исследования [98]. В то же время CRISPR-Cas9, благодаря своей популярности как удобного инструмента редактирования генома, становится все более доступным как в академической среде, так и в частном секторе [52,99]. Аналогично, технология РНК-интерференции (RNAi) доказала свою высокую применимость в ускоренной селекции и уже используется в сельском хозяйстве [100].
Разработка и совершенствование новых технологий, способствующих развитию ускоренной селекции, может проводиться на различных видах растений, однако зачастую проще разработать стандарт, тестируя новые методы на хорошо изученных видах. Именно поэтому кукуруза (*Zea mays L.*) часто используется в качестве модельного растения для испытания новых агротехнологий. Например, Сингх и др. [101] использовали кукурузу для моделирования условий освещения и температуры при ускоренной селекции, обнаружив, что она способна выдерживать более высокую интенсивность света по сравнению с другими растениями, что позволяет ей адаптироваться к уровню освещенности, характерному для условий ускоренной селекции. Кукуруза также обладает высокой эффективностью использования радиации, определяемой как количество микромолей биомассы, произведенной на моль фотосинтетически активных фотонов, поглощенных зелеными частями растения [101]. Кроме того, урожайность кукурузы за сезон сильно зависит от идеальных климатических условий, которые нарушаются под воздействием абиотических стрессов [102]. Кукуруза была использована в качестве объекта 16-линейного мультиродительского скрещивания (MAGIC) для дальнейшего изучения генетической основы взаимодействия генотипа со средой. Шестнадцать линий кукурузы исследовались в пяти умеренных зонах с контрастными климатическими условиями и различными методами управления [95]. Общие характеристики роста кукурузы, ее чувствительность к факторам среды и история использования в качестве модели в исследованиях растений [103,104] укрепили ее позиции в качестве основной культуры для тестирования и разработки технологий ускоренной селекции.
Будущие исследования могут стать пионерами в интеграции ключевых молекулярных методов, таких как геномная селекция, CRISPR-Cas9 и эпигенетическая инженерия, с растущими массивами данных из различных направлений «омиксных» исследований — от транскриптомики до протеомики — для создания системы комплексных селекционных подходов. В перспективе разработка технологии генотипирования MiMe (митоз вместо мейоза) демонстрирует потенциал для успешного внедрения в системы ускоренной селекции [52]. Дополнительные исследования апомиксиса у мутантных форм сельскохозяйственных культур также открывают путь для изучения наследования признаков в поколениях, что повышает эффективность клонального размножения растений [52]. В сочетании с оптимизированными условиями выращивания, сокращающими время генерации, эти технологии будут непрерывно улучшать способность селекционеров быстро создавать высококачественные и устойчивые сорта, способные отвечать вызовам растущего мира.
Вклад авторов
M.S. conceived the work; L.W.L., M.P. and K.W. performed the searches and wrote the manuscript, with contributions from M.S., A.T. and G.O. All authors have read and agreed to the published version of the manuscript.
Конфликт интересов
The authors declare no conflicts of interest.
Финансирование
This research received no external funding.
Примечания
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Литература
1.Lutz W., Kc S. Dimensions of global population projections: What do we know about future population trends and structures? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2010;365:2779–2791. doi: 10.1098/rstb.2010.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
2.Voss-Fels K.P., Stahl A., Hickey L.T. Q&A: Modern crop breeding for future food security. BMC Biol. 2019;17:18. doi: 10.1186/s12915-019-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
3.Kulchin Y.N., Zmeeva V.N., Subbotin E.P., Kostyanko A.A. The effect of multispectral light emitting diodes (LEDs) on the activation of morphogenic processes in cell culture of rice Oryza sativa. Defect Diffus. Forum. 2018;386:236–243. doi: 10.4028/www.scientific.net/DDF.386.236. [DOI] [Google Scholar]
4.Breseghello F., Coelho A.S.G. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.) J. Agric. Food Chem. 2013;61:8277–8286. doi: 10.1021/jf305531j. [DOI] [PubMed] [Google Scholar]
5.Anand A., Subramanian M., Kar D. Breeding techniques to dispense higher genetic gains. Front. Plant Sci. 2023;13:1076094. doi: 10.3389/fpls.2022.1076094. [DOI] [PMC free article] [PubMed] [Google Scholar]
6.Lenaerts B., Collard B.C.Y., Demont M. Review: Improving global food security through accelerated plant breeding. Plant Sci. 2019;287:110207. doi: 10.1016/j.plantsci.2019.110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
7.Raj S., Roodbar S., Brinkley C., Wolfe D.W. Food Security and Climate Change: Differences in Impacts and Adaptation Strategies for Rural Communities in the Global South and North. Front. Sustain. Food Syst. 2022;5:691191. doi: 10.3389/fsufs.2021.691191. [DOI] [Google Scholar]
8.Fischer G., Shah M., Tubiello F.N., van Velhuizen H. Socio-economic and climate change impacts on agriculture: An integrated assessment, 1990–2080. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005;360:2067–2083. doi: 10.1098/rstb.2005.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
9.Alahmad S., Dinglasan E., Leung K.M., Riaz A., Derbal N., Voss-Fels K.P., Able J.A., Bassi F.M., Christopher J., Hickey L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods. 2018;14:36. doi: 10.1186/s13007-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
10.Poulet L., Massa G.D., Morrow R.C., Bourget C.M., Wheeler R.M., Mitchell C.A. Significant reduction in energy for plant-growth lighting in space using targeted LED lighting and spectral manipulation. Life Sci. Space Res. 2014;2:43–53. doi: 10.1016/j.lssr.2014.06.002. [DOI] [Google Scholar]
11.Figueiredo M.S.N., Pereira A.M. Managing Knowledge—The Importance of Databases in the Scientific Production. Procedia Manuf. 2017;12:166–173. doi: 10.1016/j.promfg.2017.08.021. [DOI] [Google Scholar]
12.Cook T.M., Isenegger D., Dutta S., Sahab S., Kay P., Aboobucker S.I., Biswas E., Heerschap S., Nikolau B.J., Dong L., et al. Overcoming roadblocks for in vitro nurseries in plants: Induction of meiosis. Front. Plant Sci. 2023;14:1204813. doi: 10.3389/fpls.2023.1204813. [DOI] [PMC free article] [PubMed] [Google Scholar]
13.Turck F., Fornara F., Coupland G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu. Rev. Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
14.Chen M., Chory J., Fankhauser C. Light Signal Transduction in Higher Plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
15.Jiao Y., Ma L., Strickland E., Deng X.W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
16.Fang L., Ma Z., Wang Q., Nian H., Ma Q., Huang Q., Mu Y. Plant Growth and Photosynthetic Characteristics of Soybean Seedlings Under Different LED Lighting Quality Conditions. J. Plant Growth Regul. 2021;40:668–678. doi: 10.1007/s00344-020-10131-2. [DOI] [Google Scholar]
17.Ghosh S., Watson A., Gonzalez-Navarro O.E., Ramirez-Gonzalez R.H., Yanes L., Mendoza-Suárez M., Simmonds J., Wells R., Rayner T., Green P., et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018;13:2944–2963. doi: 10.1038/s41596-018-0072-z. [DOI] [PubMed] [Google Scholar]
18.Nelson J.A., Bugbee B. Economic analysis of greenhouse lighting: Light emitting diodes vs. high intensity discharge fixtures. PLoS ONE. 2014;9:e99010. doi: 10.1371/journal.pone.0099010. [DOI] [PMC free article] [PubMed] [Google Scholar]
19.Stefański P., Siedlarz-Slowacka P., Matysik P., Rybka K. Efficiency of LED lamps used in cereal crop breeding greenhouses. Int. J. Agric. Biol. Eng. 2022;15:75–83. doi: 10.25165/j.ijabe.20221502.5775. [DOI] [Google Scholar]
20.Morrow R.C. LED Lighting in Horticulture. HortScience Horts. 2008;43:1947–1950. doi: 10.21273/HORTSCI.43.7.1947. [DOI] [Google Scholar]
21.Agarwal A., Gupta S.D. Impact of Light-Emitting Diodes (LEDs) and Its Potential on Plant Growth and Development in Controlled-Environment Plant Production System. Curr. Biotechnol. 2016;5:28–43. doi: 10.2174/2211550104666151006001126. [DOI] [Google Scholar]
22.Danila E., Lucache D.D. Efficient lighting system for greenhouses; Proceedings of the 2016 International Conference and Exposition on Electrical and Power Engineering (EPE); Iasi, Romania. 20–22 October 2018; pp. 439–444. [DOI] [Google Scholar]
23.Monostori I., Heilmann M., Kocsy G., Rakszegi M., Ahres M., Altenbach S.B., Szalai G., Pál M., Toldi D., Simon-Sarkadi L., et al. LED lighting–modification of growth, metabolism, yield and flour composition in wheat by spectral quality and intensity. Front. Plant Sci. 2018;9:605–621. doi: 10.3389/fpls.2018.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
24.Singh D., Basu C., Meinhardt-Wollweber M., Roth B. LEDs for energy efficient greenhouse lighting. Renew. Sustain. Energy Rev. 2015;49:139–147. doi: 10.1016/j.rser.2015.04.117. [DOI] [Google Scholar]
25.Bantis F., Smirnakou S., Ouzounis T., Koukounaras A., Ntagkas N., Radoglou K. Current status and recent achievements in the field of horticulture with the use of light-emitting diodes (LEDs) Sci. Hortic. 2018;235:437–451. doi: 10.1016/j.scienta.2018.02.058. [DOI] [Google Scholar]
26.Hickey L.T., Hafeez A.N., Robinson H., Jackson S.A., Leal-Bertioli S.C.M., Tester M., Gao C., Godwin I.D., Hayes B.J., Wulff B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019;37:744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
27.Watson A., Ghosh S., Williams M.J., Cuddy W.S., Simmonds J., Rey M.D., Asyraf Md Hatta M., Hinchliffe A., Steed A., Reynolds D., et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants. 2018;4:23–29. doi: 10.1038/s41477-017-0083-8. [DOI] [PubMed] [Google Scholar]
28.Chiurugwi T., Kemp S., Powell W., Hickey L.T. Speed breeding orphan crops. Theor. Appl. Genet. 2019;132:607–616. doi: 10.1007/s00122-018-3202-7. [DOI] [PubMed] [Google Scholar]
29.Cha J.-K., Park H., Choi C., Kwon Y., Lee S.-M., Oh K.-W., Ko J.-M., Kwon S.-W., Lee J.-H. Acceleration of wheat breeding: Enhancing efficiency and practical application of the speed breeding system. Plant Methods. 2023;19:118. doi: 10.1186/s13007-023-01083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
30.Jähne F., Hahn V., Würschum T., Leiser W.L. Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 2020;133:2335–2342. doi: 10.1007/s00122-020-03601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
31.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
32.Richardson A.E., Barea J.M., McNeill A.M., Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009;321:305–339. doi: 10.1007/s11104-009-9895-2. [DOI] [Google Scholar]
33.Nadeem S.M., Ahmad M., Zahir Z.A., Javaid A., Ashraf M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014;32:429–448. doi: 10.1016/j.biotechadv.2013.12.005. [DOI] [PubMed] [Google Scholar]
34.Vacheron J., Desbrosses G., Bouffaud M.L., Touraine B., Moënne-Loccoz Y., Muller D., Legendre L., Wisniewski-Dyé F., Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
35.Hilou A., Zhang H., Franken P., Hause B. Do jasmonates play a role in arbuscular mycorrhiza-induced local bioprotection of Medicago truncatula against root rot disease caused by Aphanomyces euteiches? Mycorrhiza. 2014;24:45–54. doi: 10.1007/s00572-013-0513-z. [DOI] [PubMed] [Google Scholar]
36.Rybakova D., Cernava T., Köberl M., Liebminger S., Etemadi M., Berg G. Endophytes-assisted biocontrol: Novel insights in ecology and the mode of action of Paenibacillus. Plant Soil. 2016;405:125–140. doi: 10.1007/s11104-015-2526-1. [DOI] [Google Scholar]
37.Huang H.C., Erickson R.S. Effect of seed treatment with Rhizobium leguminosarum on Pythium damping-off, seedling height, root nodulation, root biomass, shoot biomass, and seed yield of pea and lentil. J. Phytopathol. 2007;155:31–37. doi: 10.1111/j.1439-0434.2006.01189.x. [DOI] [Google Scholar]
38.Faust J.E., Logan J. Daily light integral: A research review and high-resolution maps of the United States. HortScience. 2018;53:1250–1257. doi: 10.21273/HORTSCI13144-18. [DOI] [Google Scholar]
39.Fouda A., Eid A.M., Elsaied A., El-Belely E.F., Barghoth M.G., Azab E., Gobouri A.A., Hassan S.E.-D. Plant Growth-Promoting Endophytic Bacterial Community Inhabiting the Leaves of Pulicaria incisa (Lam.) DC Inherent to Arid Regions. Plants. 2021;10:76. doi: 10.3390/plants10010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
40.Fu J., Wang S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011;2:74. doi: 10.3389/fpls.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
41.Jiang C., Mithani A., Belfield E.J., Mott R., Hurst L.D., Harberd N.P. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014;24:1821–1829. doi: 10.1101/gr.177659.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
42.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
43.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
44.Garneau J.E., Dupuis M.È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
45.Chaudhuri A., Halder K., Datta A. Classification of CRISPR/Cas system and its application in tomato breeding. Theor. Appl. Genet. 2022;135:367–387. doi: 10.1007/s00122-021-03984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
46.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J. CRISPR-Cas: Adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
47.Cao H.X., Wang W., Le H.T.T., Vu G.T.H. The Power of CRISPR-Cas9-Induced Genome Editing to Speed Up Plant Breeding. Int. J. Genom. 2016;2016:5078796. doi: 10.1155/2016/5078796. [DOI] [PMC free article] [PubMed] [Google Scholar]
48.Liu J., Wang S., Wang H., Luo B., Cai Y., Li X., Zhang Y., Wang X. Rapid generation of tomato male-sterile lines with a marker use for hybrid seed production by CRISPR/Cas9 system. Mol. Breed. 2021;41:25. doi: 10.1007/s11032-021-01215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
49.Hamada H., Linghu Q., Nagira Y., Miki R., Taoka N., Imai R. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017;7:11443. doi: 10.1038/s41598-017-11936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
50.Mitter N., Worrall E.A., Robinson K.E., Li P., Jain R.G., Taochy C., Fletcher S.J., Carroll B.J., Lu G.Q., Xu Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants. 2017;3:16207. doi: 10.1038/nplants.2016.207. [DOI] [PubMed] [Google Scholar]
51.Wang M., Lu Y., Botella J.R., Mao Y., Hua K., Zhu J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant. 2017;10:1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
52.Fiaz S., Wang X., Younas A., Alharthi B., Riaz A., Ali H. Apomixis and strategies to induce apomixis to preserve hybrid vigor for multiple generations. GM Crops Food Biotechnol. Agric. Food Chain. 2021;12:57–70. doi: 10.1080/21645698.2020.1808423. [DOI] [PMC free article] [PubMed] [Google Scholar]
53.Gaafar R.M., El Shanshoury A.R., El Hisseiwy A.A., AbdAlhak M.A., Omar A.F., Abd El Wahab M.M., Nofal R.S. Induction of apomixis and fixation of heterosis in Egyptian rice Hybrid1 line using colchicine mutagenesis. Ann. Agric. Sci. 2017;62:51–60. doi: 10.1016/j.aoas.2017.03.001. [DOI] [Google Scholar]
54.Liu C., He Z., Zhang Y., Hu F., Li M., Liu Q., Huang Y., Wang J., Zhang W., Wang C., et al. Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice. Plant Commun. 2023;4:100470. doi: 10.1016/j.xplc.2022.100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
55.Aragão F.J.L., Barros L.M.G., de Sousa M.V., Grossi de Sá M.F., Almeida E.R.P., Gander E.S., Rech E.L. Expression of a methionine-rich storage albumin from the Brazil nut (Bertholletia excelsa H.B.K., Lecythidaceae) in transgenic bean plants (Phaseolus vulgaris L., Fabaceae) Genet. Mol. Biol. 1999;22:445–449. doi: 10.1590/S1415-47571999000300026. [DOI] [Google Scholar]
56.Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L., Wang Z., Zhang Z., Zheng R., Yang L., et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
57.Chen R., Gajendiran K., Wulff B.B.H. R we there yet? Advances in cloning resistance genes for engineering immunity in crop plants. Curr. Opin. Plant Biol. 2024;77:102489. doi: 10.1016/j.pbi.2023.102489. [DOI] [PubMed] [Google Scholar]
58.Johal G.S., Briggs S.P. Reductase Activity Encoded by the HM1 Disease Resistance Gene in Maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
59.Luo M., Xie L., Chakraborty S., Wang A., Matny O., Jugovich M., Kolmer J.A., Richardson T., Bhatt D., Hoque M., et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 2021;39:561–566. doi: 10.1038/s41587-020-00770-x. [DOI] [PubMed] [Google Scholar]
60.Fan X., Jiang H., Meng L., Chen J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021;22:11674. doi: 10.3390/ijms222111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
61.Pinho Morais P.P., Akdemir D., Braatz de Andrade L.R., Jannink J., Fritsche-Neto R., Borém A., Couto Alves F., Hottis Lyra D., Granato Í.S.C. Using public databases for genomic prediction of tropical maize lines. Plant Breed. 2020;139:697–707. doi: 10.1111/pbr.12827. [DOI] [Google Scholar]
62.Spindel J., Iwata H. Rice Genomics, Genetics and Breeding. Springer; Singapore: 2018. Genomic selection in rice breeding; pp. 473–496. [DOI] [Google Scholar]
63.Meuwissen T.H.E., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
64.Shamshad M., Sharma A. Next Generation Plant Breeding. InTech; Rijeka, Croatia: 2018. The Usage of Genomic Selection Strategy in Plant Breeding. [DOI] [Google Scholar]
65.Sun J., Khan M., Amir R., Gul A. Climate Change and Food Security with Emphasis on Wheat. Elsevier; Amsterdam, The Netherlands: 2020. Genomic selection in wheat breeding; pp. 321–330. [DOI] [Google Scholar]
66.Desta Z.A., Ortiz R. Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 2014;19:592–601. doi: 10.1016/j.tplants.2014.05.006. [DOI] [PubMed] [Google Scholar]
67.Alemu A., Åstrand J., Montesinos-López O.A., Isidro y Sánchez J., Fernández-Gónzalez J., Tadesse W., Vetukuri R.R., Carlsson A.S., Ceplitis A., Crossa J., et al. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant. 2024;17:552–578. doi: 10.1016/j.molp.2024.03.007. [DOI] [PubMed] [Google Scholar]
68.Diaz S., Ariza-Suarez D., Ramdeen R., Aparicio J., Arunachalam N., Hernandez C., Diaz H., Ruiz H., Piepho H.P., Raatz B. Genetic Architecture and Genomic Prediction of Cooking Time in Common Bean (Phaseolus vulgaris L.) Front. Plant Sci. 2021;11:622213. doi: 10.3389/fpls.2020.622213. [DOI] [PMC free article] [PubMed] [Google Scholar]
69.Kumar S. Epigenomics for Crop Improvement: Current Status and Future Perspectives. J. Genet. Cell Biol. 2020;3:128–134. [Google Scholar]
70.Schmid M.W., Heichinger C., Coman Schmid D., Guthörl D., Gagliardini V., Bruggmann R., Aluri S., Aquino C., Schmid B., Turnbull L.A., et al. Contribution of epigenetic variation to adaptation in Arabidopsis. Nat. Commun. 2018;9:4446. doi: 10.1038/s41467-018-06932-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
2.Voss-Fels K.P., Stahl A., Hickey L.T. Q&A: Modern crop breeding for future food security. BMC Biol. 2019;17:18. doi: 10.1186/s12915-019-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
3.Kulchin Y.N., Zmeeva V.N., Subbotin E.P., Kostyanko A.A. The effect of multispectral light emitting diodes (LEDs) on the activation of morphogenic processes in cell culture of rice Oryza sativa. Defect Diffus. Forum. 2018;386:236–243. doi: 10.4028/www.scientific.net/DDF.386.236. [DOI] [Google Scholar]
4.Breseghello F., Coelho A.S.G. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.) J. Agric. Food Chem. 2013;61:8277–8286. doi: 10.1021/jf305531j. [DOI] [PubMed] [Google Scholar]
5.Anand A., Subramanian M., Kar D. Breeding techniques to dispense higher genetic gains. Front. Plant Sci. 2023;13:1076094. doi: 10.3389/fpls.2022.1076094. [DOI] [PMC free article] [PubMed] [Google Scholar]
6.Lenaerts B., Collard B.C.Y., Demont M. Review: Improving global food security through accelerated plant breeding. Plant Sci. 2019;287:110207. doi: 10.1016/j.plantsci.2019.110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
7.Raj S., Roodbar S., Brinkley C., Wolfe D.W. Food Security and Climate Change: Differences in Impacts and Adaptation Strategies for Rural Communities in the Global South and North. Front. Sustain. Food Syst. 2022;5:691191. doi: 10.3389/fsufs.2021.691191. [DOI] [Google Scholar]
8.Fischer G., Shah M., Tubiello F.N., van Velhuizen H. Socio-economic and climate change impacts on agriculture: An integrated assessment, 1990–2080. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005;360:2067–2083. doi: 10.1098/rstb.2005.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
9.Alahmad S., Dinglasan E., Leung K.M., Riaz A., Derbal N., Voss-Fels K.P., Able J.A., Bassi F.M., Christopher J., Hickey L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods. 2018;14:36. doi: 10.1186/s13007-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
10.Poulet L., Massa G.D., Morrow R.C., Bourget C.M., Wheeler R.M., Mitchell C.A. Significant reduction in energy for plant-growth lighting in space using targeted LED lighting and spectral manipulation. Life Sci. Space Res. 2014;2:43–53. doi: 10.1016/j.lssr.2014.06.002. [DOI] [Google Scholar]
11.Figueiredo M.S.N., Pereira A.M. Managing Knowledge—The Importance of Databases in the Scientific Production. Procedia Manuf. 2017;12:166–173. doi: 10.1016/j.promfg.2017.08.021. [DOI] [Google Scholar]
12.Cook T.M., Isenegger D., Dutta S., Sahab S., Kay P., Aboobucker S.I., Biswas E., Heerschap S., Nikolau B.J., Dong L., et al. Overcoming roadblocks for in vitro nurseries in plants: Induction of meiosis. Front. Plant Sci. 2023;14:1204813. doi: 10.3389/fpls.2023.1204813. [DOI] [PMC free article] [PubMed] [Google Scholar]
13.Turck F., Fornara F., Coupland G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu. Rev. Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
14.Chen M., Chory J., Fankhauser C. Light Signal Transduction in Higher Plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
15.Jiao Y., Ma L., Strickland E., Deng X.W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
16.Fang L., Ma Z., Wang Q., Nian H., Ma Q., Huang Q., Mu Y. Plant Growth and Photosynthetic Characteristics of Soybean Seedlings Under Different LED Lighting Quality Conditions. J. Plant Growth Regul. 2021;40:668–678. doi: 10.1007/s00344-020-10131-2. [DOI] [Google Scholar]
17.Ghosh S., Watson A., Gonzalez-Navarro O.E., Ramirez-Gonzalez R.H., Yanes L., Mendoza-Suárez M., Simmonds J., Wells R., Rayner T., Green P., et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018;13:2944–2963. doi: 10.1038/s41596-018-0072-z. [DOI] [PubMed] [Google Scholar]
18.Nelson J.A., Bugbee B. Economic analysis of greenhouse lighting: Light emitting diodes vs. high intensity discharge fixtures. PLoS ONE. 2014;9:e99010. doi: 10.1371/journal.pone.0099010. [DOI] [PMC free article] [PubMed] [Google Scholar]
19.Stefański P., Siedlarz-Slowacka P., Matysik P., Rybka K. Efficiency of LED lamps used in cereal crop breeding greenhouses. Int. J. Agric. Biol. Eng. 2022;15:75–83. doi: 10.25165/j.ijabe.20221502.5775. [DOI] [Google Scholar]
20.Morrow R.C. LED Lighting in Horticulture. HortScience Horts. 2008;43:1947–1950. doi: 10.21273/HORTSCI.43.7.1947. [DOI] [Google Scholar]
21.Agarwal A., Gupta S.D. Impact of Light-Emitting Diodes (LEDs) and Its Potential on Plant Growth and Development in Controlled-Environment Plant Production System. Curr. Biotechnol. 2016;5:28–43. doi: 10.2174/2211550104666151006001126. [DOI] [Google Scholar]
22.Danila E., Lucache D.D. Efficient lighting system for greenhouses; Proceedings of the 2016 International Conference and Exposition on Electrical and Power Engineering (EPE); Iasi, Romania. 20–22 October 2018; pp. 439–444. [DOI] [Google Scholar]
23.Monostori I., Heilmann M., Kocsy G., Rakszegi M., Ahres M., Altenbach S.B., Szalai G., Pál M., Toldi D., Simon-Sarkadi L., et al. LED lighting–modification of growth, metabolism, yield and flour composition in wheat by spectral quality and intensity. Front. Plant Sci. 2018;9:605–621. doi: 10.3389/fpls.2018.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
24.Singh D., Basu C., Meinhardt-Wollweber M., Roth B. LEDs for energy efficient greenhouse lighting. Renew. Sustain. Energy Rev. 2015;49:139–147. doi: 10.1016/j.rser.2015.04.117. [DOI] [Google Scholar]
25.Bantis F., Smirnakou S., Ouzounis T., Koukounaras A., Ntagkas N., Radoglou K. Current status and recent achievements in the field of horticulture with the use of light-emitting diodes (LEDs) Sci. Hortic. 2018;235:437–451. doi: 10.1016/j.scienta.2018.02.058. [DOI] [Google Scholar]
26.Hickey L.T., Hafeez A.N., Robinson H., Jackson S.A., Leal-Bertioli S.C.M., Tester M., Gao C., Godwin I.D., Hayes B.J., Wulff B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019;37:744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
27.Watson A., Ghosh S., Williams M.J., Cuddy W.S., Simmonds J., Rey M.D., Asyraf Md Hatta M., Hinchliffe A., Steed A., Reynolds D., et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants. 2018;4:23–29. doi: 10.1038/s41477-017-0083-8. [DOI] [PubMed] [Google Scholar]
28.Chiurugwi T., Kemp S., Powell W., Hickey L.T. Speed breeding orphan crops. Theor. Appl. Genet. 2019;132:607–616. doi: 10.1007/s00122-018-3202-7. [DOI] [PubMed] [Google Scholar]
29.Cha J.-K., Park H., Choi C., Kwon Y., Lee S.-M., Oh K.-W., Ko J.-M., Kwon S.-W., Lee J.-H. Acceleration of wheat breeding: Enhancing efficiency and practical application of the speed breeding system. Plant Methods. 2023;19:118. doi: 10.1186/s13007-023-01083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
30.Jähne F., Hahn V., Würschum T., Leiser W.L. Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 2020;133:2335–2342. doi: 10.1007/s00122-020-03601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
31.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
32.Richardson A.E., Barea J.M., McNeill A.M., Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009;321:305–339. doi: 10.1007/s11104-009-9895-2. [DOI] [Google Scholar]
33.Nadeem S.M., Ahmad M., Zahir Z.A., Javaid A., Ashraf M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014;32:429–448. doi: 10.1016/j.biotechadv.2013.12.005. [DOI] [PubMed] [Google Scholar]
34.Vacheron J., Desbrosses G., Bouffaud M.L., Touraine B., Moënne-Loccoz Y., Muller D., Legendre L., Wisniewski-Dyé F., Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
35.Hilou A., Zhang H., Franken P., Hause B. Do jasmonates play a role in arbuscular mycorrhiza-induced local bioprotection of Medicago truncatula against root rot disease caused by Aphanomyces euteiches? Mycorrhiza. 2014;24:45–54. doi: 10.1007/s00572-013-0513-z. [DOI] [PubMed] [Google Scholar]
36.Rybakova D., Cernava T., Köberl M., Liebminger S., Etemadi M., Berg G. Endophytes-assisted biocontrol: Novel insights in ecology and the mode of action of Paenibacillus. Plant Soil. 2016;405:125–140. doi: 10.1007/s11104-015-2526-1. [DOI] [Google Scholar]
37.Huang H.C., Erickson R.S. Effect of seed treatment with Rhizobium leguminosarum on Pythium damping-off, seedling height, root nodulation, root biomass, shoot biomass, and seed yield of pea and lentil. J. Phytopathol. 2007;155:31–37. doi: 10.1111/j.1439-0434.2006.01189.x. [DOI] [Google Scholar]
38.Faust J.E., Logan J. Daily light integral: A research review and high-resolution maps of the United States. HortScience. 2018;53:1250–1257. doi: 10.21273/HORTSCI13144-18. [DOI] [Google Scholar]
39.Fouda A., Eid A.M., Elsaied A., El-Belely E.F., Barghoth M.G., Azab E., Gobouri A.A., Hassan S.E.-D. Plant Growth-Promoting Endophytic Bacterial Community Inhabiting the Leaves of Pulicaria incisa (Lam.) DC Inherent to Arid Regions. Plants. 2021;10:76. doi: 10.3390/plants10010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
40.Fu J., Wang S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011;2:74. doi: 10.3389/fpls.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
41.Jiang C., Mithani A., Belfield E.J., Mott R., Hurst L.D., Harberd N.P. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014;24:1821–1829. doi: 10.1101/gr.177659.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
42.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
43.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
44.Garneau J.E., Dupuis M.È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
45.Chaudhuri A., Halder K., Datta A. Classification of CRISPR/Cas system and its application in tomato breeding. Theor. Appl. Genet. 2022;135:367–387. doi: 10.1007/s00122-021-03984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
46.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J. CRISPR-Cas: Adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
47.Cao H.X., Wang W., Le H.T.T., Vu G.T.H. The Power of CRISPR-Cas9-Induced Genome Editing to Speed Up Plant Breeding. Int. J. Genom. 2016;2016:5078796. doi: 10.1155/2016/5078796. [DOI] [PMC free article] [PubMed] [Google Scholar]
48.Liu J., Wang S., Wang H., Luo B., Cai Y., Li X., Zhang Y., Wang X. Rapid generation of tomato male-sterile lines with a marker use for hybrid seed production by CRISPR/Cas9 system. Mol. Breed. 2021;41:25. doi: 10.1007/s11032-021-01215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
49.Hamada H., Linghu Q., Nagira Y., Miki R., Taoka N., Imai R. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017;7:11443. doi: 10.1038/s41598-017-11936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
50.Mitter N., Worrall E.A., Robinson K.E., Li P., Jain R.G., Taochy C., Fletcher S.J., Carroll B.J., Lu G.Q., Xu Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants. 2017;3:16207. doi: 10.1038/nplants.2016.207. [DOI] [PubMed] [Google Scholar]
51.Wang M., Lu Y., Botella J.R., Mao Y., Hua K., Zhu J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant. 2017;10:1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
52.Fiaz S., Wang X., Younas A., Alharthi B., Riaz A., Ali H. Apomixis and strategies to induce apomixis to preserve hybrid vigor for multiple generations. GM Crops Food Biotechnol. Agric. Food Chain. 2021;12:57–70. doi: 10.1080/21645698.2020.1808423. [DOI] [PMC free article] [PubMed] [Google Scholar]
53.Gaafar R.M., El Shanshoury A.R., El Hisseiwy A.A., AbdAlhak M.A., Omar A.F., Abd El Wahab M.M., Nofal R.S. Induction of apomixis and fixation of heterosis in Egyptian rice Hybrid1 line using colchicine mutagenesis. Ann. Agric. Sci. 2017;62:51–60. doi: 10.1016/j.aoas.2017.03.001. [DOI] [Google Scholar]
54.Liu C., He Z., Zhang Y., Hu F., Li M., Liu Q., Huang Y., Wang J., Zhang W., Wang C., et al. Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice. Plant Commun. 2023;4:100470. doi: 10.1016/j.xplc.2022.100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
55.Aragão F.J.L., Barros L.M.G., de Sousa M.V., Grossi de Sá M.F., Almeida E.R.P., Gander E.S., Rech E.L. Expression of a methionine-rich storage albumin from the Brazil nut (Bertholletia excelsa H.B.K., Lecythidaceae) in transgenic bean plants (Phaseolus vulgaris L., Fabaceae) Genet. Mol. Biol. 1999;22:445–449. doi: 10.1590/S1415-47571999000300026. [DOI] [Google Scholar]
56.Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L., Wang Z., Zhang Z., Zheng R., Yang L., et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
57.Chen R., Gajendiran K., Wulff B.B.H. R we there yet? Advances in cloning resistance genes for engineering immunity in crop plants. Curr. Opin. Plant Biol. 2024;77:102489. doi: 10.1016/j.pbi.2023.102489. [DOI] [PubMed] [Google Scholar]
58.Johal G.S., Briggs S.P. Reductase Activity Encoded by the HM1 Disease Resistance Gene in Maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
59.Luo M., Xie L., Chakraborty S., Wang A., Matny O., Jugovich M., Kolmer J.A., Richardson T., Bhatt D., Hoque M., et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 2021;39:561–566. doi: 10.1038/s41587-020-00770-x. [DOI] [PubMed] [Google Scholar]
60.Fan X., Jiang H., Meng L., Chen J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021;22:11674. doi: 10.3390/ijms222111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
61.Pinho Morais P.P., Akdemir D., Braatz de Andrade L.R., Jannink J., Fritsche-Neto R., Borém A., Couto Alves F., Hottis Lyra D., Granato Í.S.C. Using public databases for genomic prediction of tropical maize lines. Plant Breed. 2020;139:697–707. doi: 10.1111/pbr.12827. [DOI] [Google Scholar]
62.Spindel J., Iwata H. Rice Genomics, Genetics and Breeding. Springer; Singapore: 2018. Genomic selection in rice breeding; pp. 473–496. [DOI] [Google Scholar]
63.Meuwissen T.H.E., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
64.Shamshad M., Sharma A. Next Generation Plant Breeding. InTech; Rijeka, Croatia: 2018. The Usage of Genomic Selection Strategy in Plant Breeding. [DOI] [Google Scholar]
65.Sun J., Khan M., Amir R., Gul A. Climate Change and Food Security with Emphasis on Wheat. Elsevier; Amsterdam, The Netherlands: 2020. Genomic selection in wheat breeding; pp. 321–330. [DOI] [Google Scholar]
66.Desta Z.A., Ortiz R. Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 2014;19:592–601. doi: 10.1016/j.tplants.2014.05.006. [DOI] [PubMed] [Google Scholar]
67.Alemu A., Åstrand J., Montesinos-López O.A., Isidro y Sánchez J., Fernández-Gónzalez J., Tadesse W., Vetukuri R.R., Carlsson A.S., Ceplitis A., Crossa J., et al. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant. 2024;17:552–578. doi: 10.1016/j.molp.2024.03.007. [DOI] [PubMed] [Google Scholar]
68.Diaz S., Ariza-Suarez D., Ramdeen R., Aparicio J., Arunachalam N., Hernandez C., Diaz H., Ruiz H., Piepho H.P., Raatz B. Genetic Architecture and Genomic Prediction of Cooking Time in Common Bean (Phaseolus vulgaris L.) Front. Plant Sci. 2021;11:622213. doi: 10.3389/fpls.2020.622213. [DOI] [PMC free article] [PubMed] [Google Scholar]
69.Kumar S. Epigenomics for Crop Improvement: Current Status and Future Perspectives. J. Genet. Cell Biol. 2020;3:128–134. [Google Scholar]
70.Schmid M.W., Heichinger C., Coman Schmid D., Guthörl D., Gagliardini V., Bruggmann R., Aluri S., Aquino C., Schmid B., Turnbull L.A., et al. Contribution of epigenetic variation to adaptation in Arabidopsis. Nat. Commun. 2018;9:4446. doi: 10.1038/s41467-018-06932-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
71.Holliday R. Epigenetics: A Historical Overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
72.Kujur A., Saxena M.S., Bajaj D., Laxmi, Parida S.K. Integrated genomics and molecular breeding approaches for dissecting the complex quantitative traits in crop plants. J. Biosci. 2013;38:971–987. doi: 10.1007/s12038-013-9388-6. [DOI] [PubMed] [Google Scholar]
73.Köhler C., Springer N. Plant epigenomics—Deciphering the mechanisms of epigenetic inheritance and plasticity in plants. Genome Biol. 2017;18:132. doi: 10.1186/s13059-017-1260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
74.Kawakatsu T., Ecker J.R. Diversity and dynamics of DNA methylation: Epigenomic resources and tools for crop breeding. Breed. Sci. 2019;69:191–204. doi: 10.1270/jsbbs.19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
75.Corbin K.R., Bolt B., Rodríguez López C.M. Breeding for Beneficial Microbial Communities Using Epigenomics. Front. Microbiol. 2020;11:537237. doi: 10.3389/fmicb.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
76.Das P.R., Sherif S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020;11:946. doi: 10.3389/fpls.2020.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
77.Younis A., Siddique M.I., Kim C.-K., Lim K.-B. RNA Interference (RNAi) Induced Gene Silencing: A Promising Approach of Hi-Tech Plant Breeding. Int. J. Biol. Sci. 2014;10:1150–1158. doi: 10.7150/ijbs.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
78.Rajput M., Choudhary K., Kumar M., Vivekanand V., Chawade A., Ortiz R., Pareek N. RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants. 2021;10:1914. doi: 10.3390/plants10091914. [DOI] [PMC free article] [PubMed] [Google Scholar]
79.Abhary M., Rezk A. Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools. Springer International Publishing; Cham, Switzerland: 2015. RNAi Technology: A Potential Tool in Plant Breeding; pp. 397–427. [DOI] [Google Scholar]
80.Chaudhary D., Jeena A.S., Rohit, Gaur S., Raj R., Mishra S., Kajal, Gupta O.P., Meena M.R. Advances in RNA Interference for Plant Functional Genomics: Unveiling Traits, Mechanisms, and Future Directions. Appl. Biochem. Biotechnol. 2024:1–30. doi: 10.1007/s12010-023-04850-x. advance online publication . [DOI] [PubMed] [Google Scholar]
81.Enrique R., Siciliano F., Favaro M.A., Gerhardt N., Roeschlin R., Rigano L., Sendin L., Castagnaro A., Vojnov A., Marano M.R. Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol. J. 2011;9:394–407. doi: 10.1111/j.1467-7652.2010.00555.x. [DOI] [PubMed] [Google Scholar]
82.Jiang C.-J., Shimono M., Maeda S., Inoue H., Mori M., Hasegawa M., Sugano S., Takatsuji H. Suppression of the Rice Fatty-Acid Desaturase Gene OsSSI2 Enhances Resistance to Blast and Leaf Blight Diseases in Rice. Mol. Plant-Microbe Interact. 2009;22:820–829. doi: 10.1094/MPMI-22-7-0820. [DOI] [PubMed] [Google Scholar]
83.Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
84.Manavalan L.P., Chen X., Clarke J., Salmeron J., Nguyen H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012;63:163–175. doi: 10.1093/jxb/err258. [DOI] [PMC free article] [PubMed] [Google Scholar]
85.Wang J., Zhang L., Cao Y., Qi C., Li S., Liu L., Wang G., Mao A., Ren S., Guo Y.-D. CsATAF1 Positively Regulates Drought Stress Tolerance by an ABA-Dependent Pathway and by Promoting ROS Scavenging in Cucumber. Plant Cell Physiol. 2018;59:930–945. doi: 10.1093/pcp/pcy030. [DOI] [PubMed] [Google Scholar]
86.Guan Q., Lu X., Zeng H., Zhang Y., Zhu J. Heat stress induction of mi R 398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013;74:840–851. doi: 10.1111/tpj.12169. [DOI] [PubMed] [Google Scholar]
87.Wu J., Yu C., Huang L., Gan Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021;173:1120–1135. doi: 10.1111/ppl.13508. [DOI] [PubMed] [Google Scholar]
88.Rosa C., Kuo Y.-W., Wuriyanghan H., Falk B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018;56:581–610. doi: 10.1146/annurev-phyto-080417-050044. [DOI] [PubMed] [Google Scholar]
89.Ketumile D., Yang X., Sanchez R., Kundariya H., Rajewski J., Dweikat I.M., Mackenzie S.A. Implementation of Epigenetic Variation in Sorghum Selection and Implications for Crop Resilience Breeding. Front. Plant Sci. 2022;12:798243. doi: 10.3389/fpls.2021.798243. [DOI] [PMC free article] [PubMed] [Google Scholar]
90.Tonosaki K., Fujimoto R., Dennis E.S., Raboy V., Osabe K. Will epigenetics be a key player in crop breeding? Front. Plant Sci. 2022;13:958350. doi: 10.3389/fpls.2022.958350. [DOI] [PMC free article] [PubMed] [Google Scholar]
91.Jiang F., Doudna J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
92.Zheng X., Chen L., Xia H., Wei H., Lou Q., Li M., Li T., Luo L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017;7:39843. doi: 10.1038/srep39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
93.Ahmar S., Gill R.A., Jung K.H., Faheem A., Qasim M.U., Mubeen M., Zhou W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020;21:2590. doi: 10.3390/ijms21072590. [DOI] [PMC free article] [PubMed] [Google Scholar]
94.Teressa T., Semagehn Z., Bejiga T. Multi Environments and Genetic-Environmental Interaction (GxE) in Plant Breeding and its Challenges: A Review Article. Int. J. Res. Stud. Agric. Sci. 2021;7:11–18. doi: 10.20431/2454-6224.0704002. [DOI] [Google Scholar]
95.Hudson A.I., Odell S.G., Dubreuil P., Tixier M.-H., Praud S., Runcie D.E., Ross-Ibarra J. Analysis of genotype-by-environment interactions in a maize mapping population. G3 Genes Genomes Genet. 2022;12:jkac013. doi: 10.1093/g3journal/jkac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
96.Egea-Gilabert C., Pagnotta M.A., Tripodi P. Genotype × Environment Interactions in Crop Breeding. Agronomy. 2021;11:1644. doi: 10.3390/agronomy11081644. [DOI] [Google Scholar]
97.Budhlakoti N., Kushwaha A.K., Rai A., Chaturvedi K.K., Kumar A., Pradhan A.K., Kumar U., Kumar R.R., Juliana P., Mishra D.C., et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022;13:832153. doi: 10.3389/fgene.2022.832153. [DOI] [PMC free article] [PubMed] [Google Scholar]
98.Jannink J.-L., Lorenz A.J., Iwata H. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genom. 2010;9:166–177. doi: 10.1093/bfgp/elq001. [DOI] [PubMed] [Google Scholar]
99.Saini H., Thakur R., Gill R., Tyagi K., Goswami M. CRISPR/Cas9-gene editing approaches in plant breeding. GM Crops Food Biotechnol. Agric. Food Chain. 2023;14:1–17. doi: 10.1080/21645698.2023.2256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
100.Halder K., Chaudhuri A., Abdin M.Z., Majee M., Datta A. RNA Interference for Improving Disease Resistance in Plants and Its Relevance in This Clustered Regularly Interspaced Short Palindromic Repeats-Dominated Era in Terms of dsRNA-Based Biopesticides. Front. Plant Sci. 2022;13:885128. doi: 10.3389/fpls.2022.885128. [DOI] [PMC free article] [PubMed] [Google Scholar]
101.Singh I., Sheoran S., Kumar B., Kumar K., Rakshit S. Speed breeding in maize (Zea mays) vis-à -vis in other crops: Status and prospects. Indian J. Agric. Sci. 2021;91:1267–1273. doi: 10.56093/ijas.v91i9.116059. [DOI] [Google Scholar]
102.Farooqi M.Q.U., Nawaz G., Wani S.H., Choudhary J.R., Rana M., Sah R.P., Afzal M., Zahra Z., Ganie S.A., Razzaq A., et al. Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.) Front. Plant Sci. 2022;13:965878. doi: 10.3389/fpls.2022.965878. [DOI] [PMC free article] [PubMed] [Google Scholar]
103.Butrón A., Santiago R., Gowda M. Editorial: Model organisms in plant science: Maize. Front. Plant Sci. 2023;14:1147857. doi: 10.3389/fpls.2023.1147857. [DOI] [PMC free article] [PubMed] [Google Scholar]
104.Hake S., Ross-Ibarra J. Genetic, evolutionary and plant breeding insights from the domestication of maize. eLife. 2015;4:e05861. doi: 10.7554/eLife.05861. [DOI] [PMC free article] [PubMed] [Google Scholar]
72.Kujur A., Saxena M.S., Bajaj D., Laxmi, Parida S.K. Integrated genomics and molecular breeding approaches for dissecting the complex quantitative traits in crop plants. J. Biosci. 2013;38:971–987. doi: 10.1007/s12038-013-9388-6. [DOI] [PubMed] [Google Scholar]
73.Köhler C., Springer N. Plant epigenomics—Deciphering the mechanisms of epigenetic inheritance and plasticity in plants. Genome Biol. 2017;18:132. doi: 10.1186/s13059-017-1260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
74.Kawakatsu T., Ecker J.R. Diversity and dynamics of DNA methylation: Epigenomic resources and tools for crop breeding. Breed. Sci. 2019;69:191–204. doi: 10.1270/jsbbs.19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
75.Corbin K.R., Bolt B., Rodríguez López C.M. Breeding for Beneficial Microbial Communities Using Epigenomics. Front. Microbiol. 2020;11:537237. doi: 10.3389/fmicb.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
76.Das P.R., Sherif S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020;11:946. doi: 10.3389/fpls.2020.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
77.Younis A., Siddique M.I., Kim C.-K., Lim K.-B. RNA Interference (RNAi) Induced Gene Silencing: A Promising Approach of Hi-Tech Plant Breeding. Int. J. Biol. Sci. 2014;10:1150–1158. doi: 10.7150/ijbs.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
78.Rajput M., Choudhary K., Kumar M., Vivekanand V., Chawade A., Ortiz R., Pareek N. RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants. 2021;10:1914. doi: 10.3390/plants10091914. [DOI] [PMC free article] [PubMed] [Google Scholar]
79.Abhary M., Rezk A. Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools. Springer International Publishing; Cham, Switzerland: 2015. RNAi Technology: A Potential Tool in Plant Breeding; pp. 397–427. [DOI] [Google Scholar]
80.Chaudhary D., Jeena A.S., Rohit, Gaur S., Raj R., Mishra S., Kajal, Gupta O.P., Meena M.R. Advances in RNA Interference for Plant Functional Genomics: Unveiling Traits, Mechanisms, and Future Directions. Appl. Biochem. Biotechnol. 2024:1–30. doi: 10.1007/s12010-023-04850-x. advance online publication . [DOI] [PubMed] [Google Scholar]
81.Enrique R., Siciliano F., Favaro M.A., Gerhardt N., Roeschlin R., Rigano L., Sendin L., Castagnaro A., Vojnov A., Marano M.R. Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol. J. 2011;9:394–407. doi: 10.1111/j.1467-7652.2010.00555.x. [DOI] [PubMed] [Google Scholar]
82.Jiang C.-J., Shimono M., Maeda S., Inoue H., Mori M., Hasegawa M., Sugano S., Takatsuji H. Suppression of the Rice Fatty-Acid Desaturase Gene OsSSI2 Enhances Resistance to Blast and Leaf Blight Diseases in Rice. Mol. Plant-Microbe Interact. 2009;22:820–829. doi: 10.1094/MPMI-22-7-0820. [DOI] [PubMed] [Google Scholar]
83.Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
84.Manavalan L.P., Chen X., Clarke J., Salmeron J., Nguyen H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012;63:163–175. doi: 10.1093/jxb/err258. [DOI] [PMC free article] [PubMed] [Google Scholar]
85.Wang J., Zhang L., Cao Y., Qi C., Li S., Liu L., Wang G., Mao A., Ren S., Guo Y.-D. CsATAF1 Positively Regulates Drought Stress Tolerance by an ABA-Dependent Pathway and by Promoting ROS Scavenging in Cucumber. Plant Cell Physiol. 2018;59:930–945. doi: 10.1093/pcp/pcy030. [DOI] [PubMed] [Google Scholar]
86.Guan Q., Lu X., Zeng H., Zhang Y., Zhu J. Heat stress induction of mi R 398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013;74:840–851. doi: 10.1111/tpj.12169. [DOI] [PubMed] [Google Scholar]
87.Wu J., Yu C., Huang L., Gan Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021;173:1120–1135. doi: 10.1111/ppl.13508. [DOI] [PubMed] [Google Scholar]
88.Rosa C., Kuo Y.-W., Wuriyanghan H., Falk B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018;56:581–610. doi: 10.1146/annurev-phyto-080417-050044. [DOI] [PubMed] [Google Scholar]
89.Ketumile D., Yang X., Sanchez R., Kundariya H., Rajewski J., Dweikat I.M., Mackenzie S.A. Implementation of Epigenetic Variation in Sorghum Selection and Implications for Crop Resilience Breeding. Front. Plant Sci. 2022;12:798243. doi: 10.3389/fpls.2021.798243. [DOI] [PMC free article] [PubMed] [Google Scholar]
90.Tonosaki K., Fujimoto R., Dennis E.S., Raboy V., Osabe K. Will epigenetics be a key player in crop breeding? Front. Plant Sci. 2022;13:958350. doi: 10.3389/fpls.2022.958350. [DOI] [PMC free article] [PubMed] [Google Scholar]
91.Jiang F., Doudna J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
92.Zheng X., Chen L., Xia H., Wei H., Lou Q., Li M., Li T., Luo L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017;7:39843. doi: 10.1038/srep39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
93.Ahmar S., Gill R.A., Jung K.H., Faheem A., Qasim M.U., Mubeen M., Zhou W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020;21:2590. doi: 10.3390/ijms21072590. [DOI] [PMC free article] [PubMed] [Google Scholar]
94.Teressa T., Semagehn Z., Bejiga T. Multi Environments and Genetic-Environmental Interaction (GxE) in Plant Breeding and its Challenges: A Review Article. Int. J. Res. Stud. Agric. Sci. 2021;7:11–18. doi: 10.20431/2454-6224.0704002. [DOI] [Google Scholar]
95.Hudson A.I., Odell S.G., Dubreuil P., Tixier M.-H., Praud S., Runcie D.E., Ross-Ibarra J. Analysis of genotype-by-environment interactions in a maize mapping population. G3 Genes Genomes Genet. 2022;12:jkac013. doi: 10.1093/g3journal/jkac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
96.Egea-Gilabert C., Pagnotta M.A., Tripodi P. Genotype × Environment Interactions in Crop Breeding. Agronomy. 2021;11:1644. doi: 10.3390/agronomy11081644. [DOI] [Google Scholar]
97.Budhlakoti N., Kushwaha A.K., Rai A., Chaturvedi K.K., Kumar A., Pradhan A.K., Kumar U., Kumar R.R., Juliana P., Mishra D.C., et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022;13:832153. doi: 10.3389/fgene.2022.832153. [DOI] [PMC free article] [PubMed] [Google Scholar]
98.Jannink J.-L., Lorenz A.J., Iwata H. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genom. 2010;9:166–177. doi: 10.1093/bfgp/elq001. [DOI] [PubMed] [Google Scholar]
99.Saini H., Thakur R., Gill R., Tyagi K., Goswami M. CRISPR/Cas9-gene editing approaches in plant breeding. GM Crops Food Biotechnol. Agric. Food Chain. 2023;14:1–17. doi: 10.1080/21645698.2023.2256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
100.Halder K., Chaudhuri A., Abdin M.Z., Majee M., Datta A. RNA Interference for Improving Disease Resistance in Plants and Its Relevance in This Clustered Regularly Interspaced Short Palindromic Repeats-Dominated Era in Terms of dsRNA-Based Biopesticides. Front. Plant Sci. 2022;13:885128. doi: 10.3389/fpls.2022.885128. [DOI] [PMC free article] [PubMed] [Google Scholar]
101.Singh I., Sheoran S., Kumar B., Kumar K., Rakshit S. Speed breeding in maize (Zea mays) vis-à -vis in other crops: Status and prospects. Indian J. Agric. Sci. 2021;91:1267–1273. doi: 10.56093/ijas.v91i9.116059. [DOI] [Google Scholar]
102.Farooqi M.Q.U., Nawaz G., Wani S.H., Choudhary J.R., Rana M., Sah R.P., Afzal M., Zahra Z., Ganie S.A., Razzaq A., et al. Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.) Front. Plant Sci. 2022;13:965878. doi: 10.3389/fpls.2022.965878. [DOI] [PMC free article] [PubMed] [Google Scholar]
103.Butrón A., Santiago R., Gowda M. Editorial: Model organisms in plant science: Maize. Front. Plant Sci. 2023;14:1147857. doi: 10.3389/fpls.2023.1147857. [DOI] [PMC free article] [PubMed] [Google Scholar]
104.Hake S., Ross-Ibarra J. Genetic, evolutionary and plant breeding insights from the domestication of maize. eLife. 2015;4:e05861. doi: 10.7554/eLife.05861. [DOI] [PMC free article] [PubMed] [Google Scholar]
